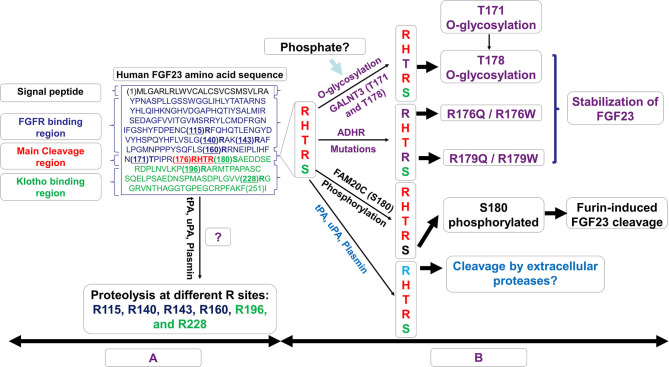
Figure 2.
Schematic representation of the possible fates of FGF23. In (A), the amino acid sequence of human FGF23 is shown with its signal peptide, FGFR binding region, the RXXR motif/cleavage sequence, and the Klotho binding region. In (B), the possible fates of mature FGF23 after the loss of its signal peptide sequence is displayed. The O-glycosylation of T178, a process controlled by N-acetylgalactosaminyltransferase 3 (GALNT3) and which requires a precedent glycosylation at T171, stabilizes and prevents FGF23 cleavage. This process may also be stimulated by increased levels of phosphate which induces GALNT3 and the glycosylation. Genetic modifications such as autosomal dominant hypophosphataemic rickets (ADHR) mutations R176Q, R176W, R179Q, and R179W stabilize FGF23 and counteract its natural cleavage. Phosphorylation at position S180 by FAM20C increases FGF23 cleavage controlled by the furin/subtilisin-like proprotein convertase. FGF23 proteolysis may occur at R176 by extracellular proteases of the plasminogen activation system, tissue-type PA (tPA), urokinase-type PA (uPA), and plasmin. This proteolysis could potentially occur at the RXXR motif (R176) as well as at different arginine residue sites at R115, R140, R143, R160, R196, and R228.