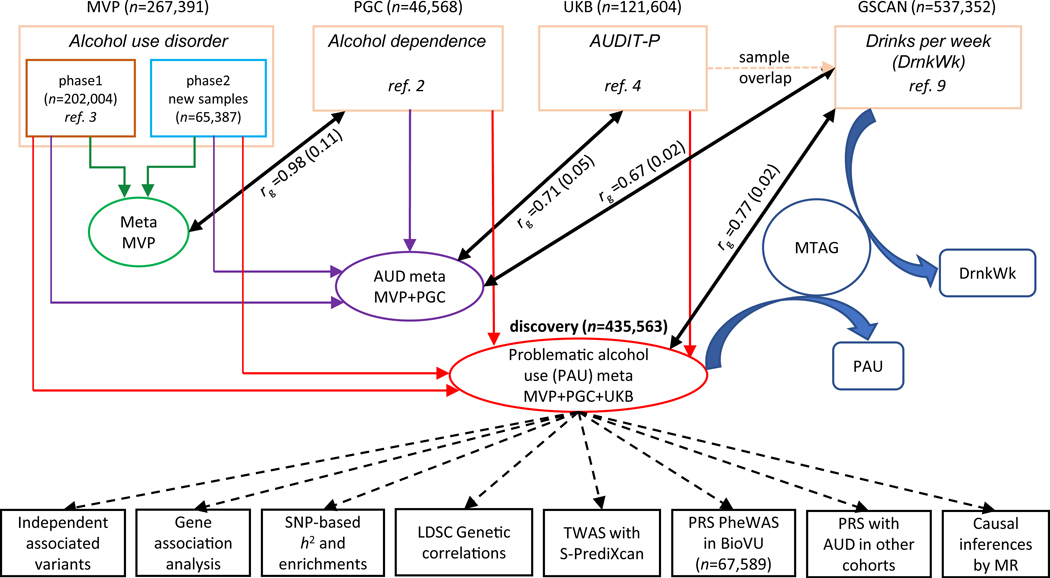
Figure 1. Overview of the analysis.
The four datasets that were meta-analyzed as the discovery sample for problematic alcohol use (PAU) included MVP phase1, MVP phase2, PGC, and UK Biobank (UKB). MVP phase1 and phase2 were meta-analyzed, and the result was used to test the genetic correlation with PGC alcohol dependence. An intermediary meta-analysis (AUD meta) combining MVP phase1, phase2, and PGC was then conducted to measure the genetic correlation with UKB AUDIT-P. Due to the sample overlap between UKB and GSCAN, we used the AUD (intermediary) meta-analysis for Mendelian Randomization (MR) analysis rather than the PAU (i.e., from the final) meta-analysis. MTAG, which used the summary data from PAU and DrnkWk (drinks per week) in GSCAN (without 23andMe samples, as those data were not available) as input to increase the power for each trait without introducing bias from sample overlap, returned summary results for PAU and DrnkWk separately.