FIG 1.
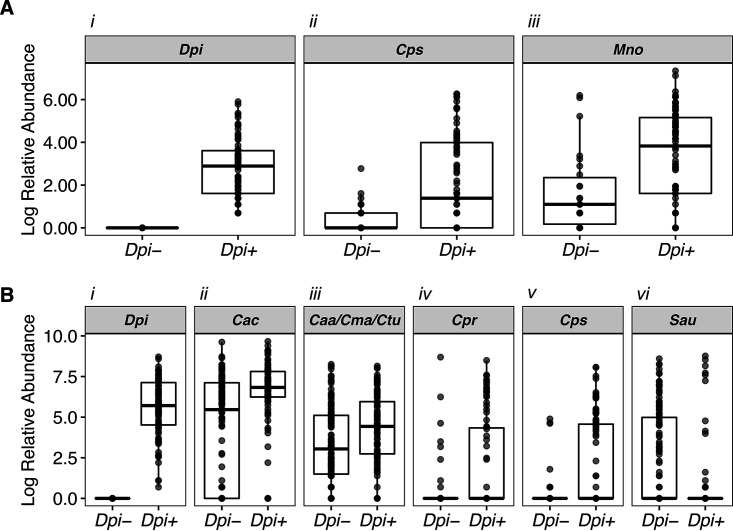
Individual nasal Corynebacterium species exhibit increased differential relative abundances in the presence of D. pigrum in human nostril microbiota. We used ANCOM to compare the species/supraspecies-level compositions of 16S rRNA gene nostril data sets from (A) 99 children ages 6 to 78 months and (B) 210 adults where D. pigrum was either absent (Dpi−) or present (Dpi+) on the basis of 16S rRNA gene sequencing data. Plots show only the taxa identified as statistically significant (sig = 0.05) after correction for multiple testing within ANCOM. The dark bar represents the median; lower and upper hinges correspond to the first and third quartiles. Each gray dot represents the value for a sample, and multiple overlapping dots appear black. Dpi = Dolosigranulum pigrum, Cac = Corynebacterium accolens, Caa/Cma/Ctu = supraspecies Corynebacterium accolens_macginleyi_tuberculostearicum, Cpr = Corynebacterium propinquum, Cps = Corynebacterium pseudodiphtheriticum, Mno = Moraxella nonliquefaciens. Only three species and one supraspecies of Corynebacterium from among the larger number of Corynebacterium supraspecies/species present in each data set met the significance threshold. Specifically, in the adult nostril data set, there were 21 species and 5 supraspecies groupings of Corynebacterium in addition to the reads of Corynebacterium that were nonassigned (NA) at the species level. These data were previously published (see Table S7 in reference 41). In the pediatric data set, there were 16 species of Corynebacterium in addition to those that were nonassigned among the species-level Corynebacterium reads (see Table S2). The Log relative abundance numerical data represented in this figure are available in Table S1.