Abstract
In this study, third-generation full-length (FL) transcriptome sequencing was performed of loquat using single-molecule real-time(SMRT) sequencing from the pooled cDNA of embryos of young loquat fruit under different low temperatures (three biological replicates for treatments of 1°C, -1°C, and -3°C, for 12 h or 24 h) and the control group(three biological replicates for treatments of room temperature), Illumina sequencing was used to correct FL transcriptome sequences. A total of 3 PacBio Iso-Seq libraries (1–2 kb, 2–3 kb and 3–6 kb) and 21 Illumina transcriptome libraries were constructed, a total of 13.41 Gb of clean reads were generated, which included 215,636 reads of insert (ROIs) and 121,654 FL, non-chimaric (FLNC) reads. Transcript clustering analysis of the FLNC reads revealed 76,586 consensus isoforms, and a total of 12,520 high-quality transcript sequences corrected with non-FL sequences were used for subsequent analysis. After the redundant reads were removed, 38,435 transcripts were obtained. A total of 27,905 coding DNA sequences (CDSs) were identified, and 407 long non-coding RNAs (lncRNAs) were ultimately predicted. Additionally, 24,832 simple sequence repeats (SSRs) were identified, and a total of 1,295 alternative splicing (AS) events were predicted. Furthermore, 37,993 transcripts were annotated in eight functional databases. This is the first study to perform SMRT sequencing of the FL transcriptome of loquat. The obtained transcriptomic data are conducive for further exploration of the mechanism of loquat freezing injury and thus serve as an important theoretical basis for generating new loquat material and for identifying new ways to improve loquat cold resistance.
Introduction
Loquat (Eriobotrya japonica Lindl) originated in China and has been cultivated for 2100 years. Owing to its economic and ecological attributes, loquat is an important perennial fruit crop species and is cultivated largely between the N 35° and S 35° latitudes worldwide [1–2]. Loquat blossoms in late autumn or early winter, and young fruits are vulnerable to freezing injury [3–5]. In 2016, 90% of the loquat planting area in China experienced freezing, with almost no material harvested. Freezing injury has severely jeopardized the economic benefits of farmers and has become a major restricting factor for sustainable development in many production areas worldwide. Current research on loquat has mainly focused on cell genetics [6, 7], physiology and biochemistry [8, 9], molecular markers [10], molecular clones [11, 12], etc. Several transcriptome studies in loquat focused on flower bud differentiation [13], fruit development and ripening [14, 15], and postharvest storage [16], research on transcriptome in cold stress of loquat is limited [17], little is known about its cold tolerance mechanisms. Previous studies have been performed using second generation sequencing technology, and many unigenes have been obtained, however, transcriptomic sequences using second generation sequencing technology may be misassembled without a high-quality genome sequence or full-length (FL) transcriptomic sequences available as a reference [18]. To date, FL transcriptomic data are scarce. In addition, Loquat is a non-model plant species with high heterozygosity, and a loquat reference genome is still lacking, which has limited molecular biological research of this species.
In recent years, third-generation sequencing technology has been successfully applied to functional genomics research of sweet potato [19], Populus [20], sorghum [21], corn [22], and cotton [23], among others. Compared with second-generation sequencing technology,third-generation sequencing technology not only has advantages that include handling a large volume of data and the ability to read long sequences and FL gene transcripts, but it is also greatly more accurate in terms of gene functional annotation without sequence splicing and assembly [24].
In the present study, The FL transcriptome of embryos of young loquat fruit under low-temperature stress was obtained by single-molecule real-time (SMRT) sequencing. This work will facilitate future research on identifying functional genes and analysing molecular mechanisms related to the cold stress response of loquat.
Materials and methods
Plant materials and treatments
Two-year-old grafted Ninghaibai loquat plants that were growing in pots and that had already produced fruit (with a diameter of approximately 1.5 cm) were used as the experimental materials, and the growth status of the plants was as uniform as possible. The plants were subjected to three different temperatures, 1°C, -1°C, and -3°C, for 12 h or 24 h separately after being subjected to a gradient of cooling at a rate of 4°C/h. The treatments were applied in a low-temperature plant incubator with 60% relative humidity, a 3000 lx light intensity, and a 12-h/12-h light/dark cycle. The plants were then removed and incubated at room temperature for 6 h to recover, after which the embryos of young loquat fruit were collected, immediately frozen in liquid nitrogen and stored at -80°C.
Plants that had been growing at room temperature were used as controls. Each treatment involved three biological replications. A total of 21 samples of embryos of young loquat fruit (three biological replicates for treatments of 1°C, -1°C, and -3°C, for 12 h or 24 h, including the control group) were collected for the following experiments.
RNA extraction and quantification
Total RNA was extracted with the RNAprep Pure Plant Kit (TIANGEN, Cat. No. DP441) following the manufacturer’s protocol. The samples were quantified as follows. The purity and concentration of RNA were first measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Rockland, DE, USA) according to their OD260/280 value, after which the RNA integrity was assessed using an RNA Nano 6000 Assay Kit in conjunction with an Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The RNA degradation and contamination were measured on 1% agarose gels. Only total RNAs with a RIN score ≥8.0 were used to construct cDNA libraries for SMRT or Illumina sequencing.
PacBio Iso-Seq library preparation and sequencing
After the RNA quality was verified, libraries were constructed. mRNA was purifed from 3μg of mixed total RNA of 21 samples of embryos of young loquat fruit for SMRT library preparation and sequencing. The instruments used include a SMARTer™ PCR cDNA Synthesis Kit (Clontech, CA, USA)and BluePippin® Size Selection System (Sage Science, Beverly, MA, USA). The SMARTer™ PCR cDNA Synthesis Kit (Clontech, CA, USA) was used for synthesizing FL cDNA, the generated cDNAs were then reamplified via PCR. The remaining overhangs were converted to blunt ends by exonuclease/polymerase activities. After adenylation of the 3′ ends of the DNA fragments, NEBNext Adaptors with a hairpin loop structure were ligated in preparation for hybridization. The BluePippin® Size Selection System was used for size selection(1–2 kb, 2–3 kb and 3–6 kb) to bulid 3 libraries.
The quality of the libraries was assessed using an Agilent Bioanalyzer 2100 system, and SMRT sequencing was performed using a Pacific Biosciences real-time sequencer in conjunction with C2 sequencing reagent.
Illumina transcriptome library preparation and sequencing
21 second-generation-sequencing cDNA libraries of embryos of young loquat fruit (three biological replicates for treatments of 1°C, -1°C, and -3°C, for 12 h or 24 h, including the control group) were constructed respectively using a NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, Beverly, MA, USA) according to the manufacturer’s protocol. Briefly, mRNA was purified from 5μg of total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under high temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First-strand cDNA was synthesized using random hexamer primers and M-MuLV Reverse Transcriptase (RNase H-). Second-strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. The remaining overhangs were converted to blunt ends via exonuclease/polymerase activities. After poly-adenylation of the 3’ ends of the DNA fragments, NEBNext adaptors with hairpin loop structures were ligated in preparation for hybridization. An AMPure XP system (Beckman Coulter, Beverly, USA) was used to select cDNA fragments that were 200–250 bp in length. Afterward, 3 μl of USER Enzyme (NEB, USA) together with size-selected, adaptor-ligated cDNA was incubated at 37°C for 15 min and again at 95°C for 5 min. PCR was then performed with Phusion High-Fidelity DNA Polymerase, universal PCR primers, and Index (X) Primer. The PCR products were ultimately purified (AMPure XP system), and the library quality was assessed using the Agilent 2100 system. The qualified libraries were pair-end sequenced on an Illumina HiSeq 2500 (Illumina, San Diego, CA, USA) system.
Error correction and quality control of SMRT reads
Raw data (raw reads) in fastq format were first processed using in-house Perl scripts. Raw SMRT sequencing reads were processed by removing polymerase reads that were <50 bp and had a accuracy <0.8, resulting in subreads. The joined subreads were disconnected, and joint sequences that were <50 bp were removed, resulting in clean data. The obtained clean reads were processed into error-corrected reads of inserts (ROIs) with parameters including full passes ≥0 and a sequence accuracy ≥0.8. Then, full-length, non-chimeric (FLNC) transcripts were determined by searching for poly-A tail signals and the 5’ and 3’ cDNA primers within the ROIs. Iterative clustering for error correction (ICE) [25] was used to obtain consensus isoforms, and FL consensus sequences from ICE were polished using Quiver. High-quality FL transcripts were classified as those with a post-correction accuracy criterion surpassing 99%. Any redundancy in high-quality, FL transcripts was removed by CD-HIT [26], and the integrity of the transcriptome was evaluated without redundancy by BUSCO [27].
Alternative splicing (AS) detection
We subjected Iso-Seq™ data directly to an all-vs-all BLAST analysis [28], with high identity settings. The BLAST alignments that met all the criteria were considered products of candidate AS events [29]. There should be two high-scoring segment pairs (HSPs) in the alignment: two HSPs had the same forward/reverse direction, and within the same alignment, one sequence should be continuous, or with a small "overlap" size (smaller than 5 bp); the other sequence should be distinct to show an "AS gap", and the continuous sequence should align to the distinct sequence almost completely. The AS gap should be larger than 100 bp and at least 100 bp away from the 3'/5' end.
Simple sequence repeat (SSR) detection
Transcripts >500 bp were selected for SSR analysis using the MIcroSAtellite identification tool (MISA; http://pgrc.ipk-gatersleben.de/misa/http://pgrc.ipk-gatersleben.de/misa/). MISA was used to identify seven SSR types, namely, mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide, hexanucleotide, and compound SSRs, by analysing transcript sequences.
Prediction of coding DNA sequences (CDSs)
The CDSs and corresponding amino acid sequences within the transcript sequences were predicted using TransDecoder (https://github.com/TransDecoder/TransDecoder/releases). TransDecoder was used to identify candidate protein-coding regions based on the open reading frame (ORF) length, log-likelihood score, nucleotide composition, and (optional) Pfam domain content [30].
Long non-coding RNA (lncRNA) prediction
Putative protein-coding RNAs were filtered and removed using the following minimum length and exon number thresholds. Transcripts that were longer than 200 nt and that had more than two exons were selected as lncRNA candidates and further screened using the Coding Potential Calculator (CPC) [31]/Coding-Non-Coding Index (CNCI/Coding Potential Assessment Tool (CPAT) [32]/Pfam database, which has the power to distinguish protein-coding genes from non-coding genes.
Functional annotation of transcripts and analysis of transcription factors (TFs)
The non-redundant transcript sequences obtained were mapped to eight different databases to obtain annotation information associated with the transcripts. These databases included the non-redundant (NR) [33], Swiss-Prot [34], Gene Ontology (GO; http://www.geneontology.org) [35], Clusters of Orthologous Groups of proteins (COG; http://www.ncbi.nlm.nih.gov/COG) [36], euKaryotic Orthologous Groups (KOG) [37], Pfam (http://pfam.janelia.org/) [38], evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG; http://eggnog.embl.de), and Kyoto Encyclopaedia of Genes and Genomes (KEGG, http://www.genome.ad.jp/kegg/) databases [39].
Finally, TFs were predicted using iTAK [40] predictive software.
Results
SMRT- and Illumina-based RNA sequencing and error correction
A total of 13.41 Gb of clean data were generated via Pacific Biosciences SMRT sequencing technology. Based on the conditions of full passes ≥0 and a quality >0.8, a total of 215,636 reads of inserts (ROIs)were obtained (Table 1), and 121,654 full-length non-chimeric (FLNC) sequences were identified (Table 2). In total, 76,586 consensus isoforms were obtained by iterative clustering for error correction(ICE) (Table 3). After error correction with second-generation sequencing short reads was performed, a total of 38,435 non-redundant transcripts with an average length of 2607bp were obtained, including 12,520 high-quality transcripts. All the raw data were deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA623262 and are available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA623262.
Table 1. ROI statistics.
| Samples | cDNA size | Reads of insert | Total read bases of insert | Mean read length of insert | Mean read quality of insert | Mean number of passes |
|---|---|---|---|---|---|---|
| F01 | 1–2 kb | 60768 | 112445632 | 1850 | 0.96 | 3 |
| F01 | 2–3 kb | 82481 | 214455943 | 2600 | 0.95 | 7 |
| F01 | 3–6 kb | 72387 | 244539490 | 3378 | 0.94 | 5 |
| F01 | All | 215636 | 571441065 | 2609.33 | 0.95 | 5 |
Table 2. FL sequence statistics.
| Samples | cDNA size | Reads of insert | Number of 5’ reads | Number of 3’ reads | Number of poly-A reads | Number of filtered short reads | Number of non-full-length reads | Number of full-length reads | Number of full-length non-chimeric reads | Number of full-length non-chimeric bases | Average length of full-length non-chimeric reads | Full-length percentage (FL%) | Artificial concatemers (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F01 | 1–2 kb | 60768 | 29868 | 34965 | 35769 | 5344 | 30620 | 24804 | 24719 | 35410268 | 1432 | 40.82 | 0.34 |
| F01 | 2–3 kb | 82481 | 67290 | 69380 | 68092 | 939 | 22155 | 59387 | 59167 | 151411494 | 2559 | 72.00 | 0.37 |
| F01 | 3–6 kb | 72387 | 48656 | 51394 | 49502 | 810 | 33665 | 37912 | 37768 | 130361660 | 3451 | 52.37 | 0.38 |
| F01 | All | 215636 | 145814 | 155739 | 153363 | 7093 | 86440 | 122103 | 121654 | 317183422 | 2607 | 56.62 | 0.36 |
Table 3. ICE clustering results statistics.
| Samples | Number of consensus isoforms | Average read length of consensus isoforms | Number of polished high-quality isoforms | Number of polished low-quality isoforms | Percent of polished high-quality isoforms (%) |
|---|---|---|---|---|---|
| F01 | 76586 | 2655 | 12520 | 63997 | 16.35 |
Predictions of CDSs, lncRNAs, and SSRs
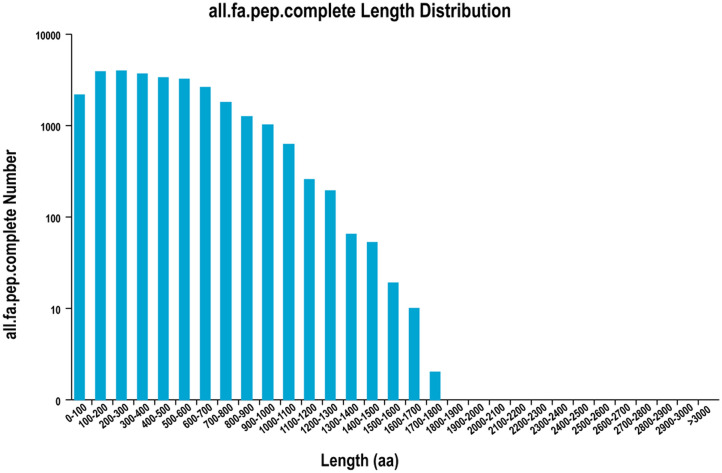
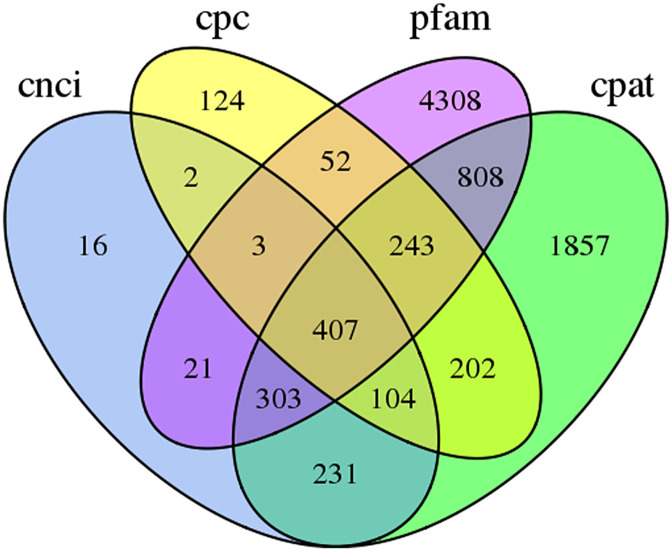
A total of 1,295 alternative splicing (AS) sequences were obtained. There were 37,230 ORFs that included 27,905 CDSs identified by TransDecoder, the distribution of the coding sequence lengths of complete ORFs is shown in Fig 1. Four computational approaches (CPC analysis, CNCI analysis, Pfam protein domain analysis, CPAT analysis) were used to screen the transcripts that encode coding proteins (Fig 2), and 407 lncRNAs were predicted. Transcripts that were >500 bp were selected for SSR analysis using MISA. In total, 24,832 SSRs were identified, including 5,317 sequences containing more than 1 SSR and 3,536 SSRs present in compound formation. Moreover, SSRs consisting of one to six (mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides) tandem repeats were identified, Mono-nucleotid repeats (12,230) were the most abundant, followed by di-nucleotid repeats (8857), tri-nucleotid repeats (3327), tetra-nucleotide repeats (254), hexa-nucleotide repeats (95) and penta-nucleotide repeats (69) (Table 4).
Fig 1. Distribution of the lengths of CDSs within complete ORFs.
The x-axis represents the coding sequence length; the y-axis represents the number of predicted ORFs.
Fig 2. Venn diagram of the number of lncRNAs predicted by CPC, CNCI, CPAT, and Pfam protein structure domain analyses.
Table 4. Statistical analysis of the SSR results.
| Item | Total number of sequences examined | Total size of examined sequences (bp) | Total number of identified SSRs | Number of SSR containing sequences | Number of sequences containing more than 1 SSR | Number of SSRs present in compound formation | Mono- nucleotides | Di- nucleotides | Tri- nucleotides | Tetra- nucleotides | Penta- nucleotides | Hexa- nucleotides |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Numbers | 38412 | 99784911 | 24832 | 16390 | 5317 | 3536 | 12230 | 8857 | 3327 | 254 | 69 | 95 |
Transcript functional annotation and sorting of transcription factors
In total, 37,993 transcripts were annotated in eight databases (Table 5). Among these transcripts, 37,908 were annotated in the NCBI NR database, 16,261 were annotated in the COG database, 22,732 in the GO database, 16,507 in the KEGG database, 24,787 in the KOG database, 31,494 in the Pfam database, 28,599 in the Swiss-Prot database, and 37,074 in the eggNOG database.
Table 5. Numbers of annotated transcripts in the publicly available databases.
| Annotated databases | Transcript number |
|---|---|
| Clusters of Orthologous Groups(COG) | 16261 |
| Gene Ontology(GO) | 22732 |
| Kyoto Encyclopaedia of Genes and Genomes(KEGG) | 16507 |
| Eukaryotic Ortholog Groups(KOG) | 24787 |
| protein family(Pfam) | 31494 |
| The Swiss-Prot Protein Knowledgebase(Swiss-Prot) | 28599 |
| Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups(eggNOG) | 37074 |
| NCBI Non-Redundant Protein Database(NR) | 37908 |
| All annotated | 37993 |
| All analysed | 38435 |
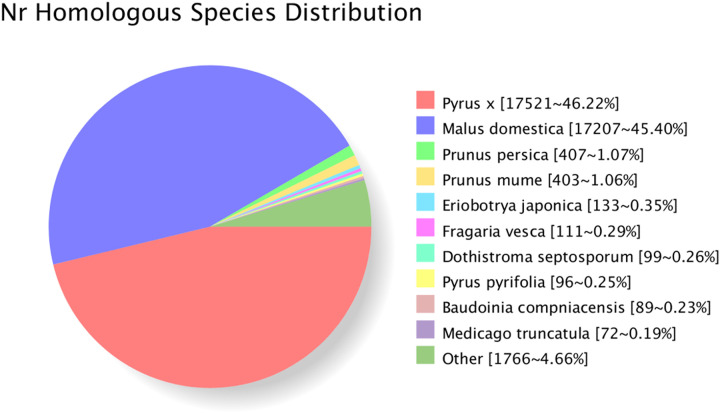
NR contains protein data from the Swiss-Prot, Protein Information Resource, Protein Research Foundation, Protein Data Bank, GenBank, and RefSeq databases;it is a non-redundant protein database housed within the NCBI. The non-redundant transcripts were compared to those in the NR database,the results showed that 46.22% of sequences were aligned to Pyrus x, followed by Malus domestica(45.40%), only 0.35% of sequences were aligned to loquat itself (Fig 3).
Fig 3. NR annotation of loquat transcripts.
GOanalysis indicated that 22,732 transcripts enriched in the pathways related to biological processes, cellular components, and molecular functions. A large number of transcripts in ‘‘cellular components” were mainly involved in cell part, cell, organelle, membrance, membrane part, and macromolecular complex. The category ‘‘molecular functions” mainly consisted of transcripts involved in catalytic activity, binding and transporter activity. The category ‘‘biological process” mainly consisted of transcripts involved in metabolic process, cellular process, single-organism process,biological regulation, localization, responses to stimulus, and cellular component organization or biogenesis (Fig 4).
Fig 4. GO functional annotation of loquat transcripts.
The x-axis represents GO categories, the y-axis (right) represents the number of transcripts, and the y-axis (left) represents the percentage of transcripts.
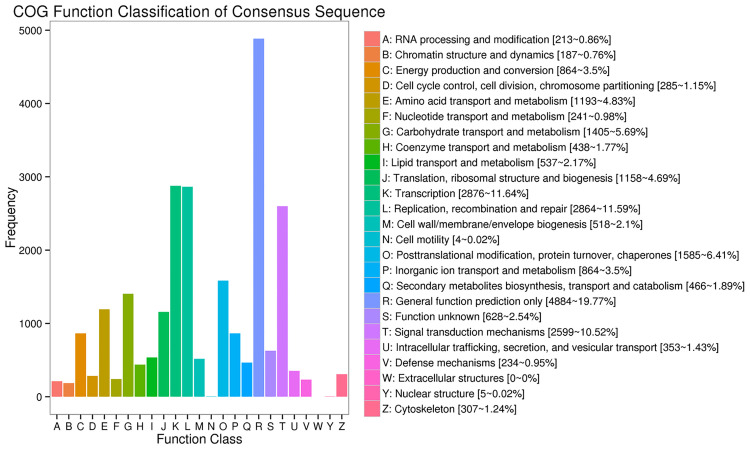
In the COG database, we found that the R function (general function prediction only) had the largest number, followed by the K function (transcription), L function (replication, recombination, and repair), and T function (signal transduction mechanisms) (Fig 5).
Fig 5. COG annotations of loquat transcripts.
The x-axis represents the COG categories, and the y-axis represents the number of transcripts.
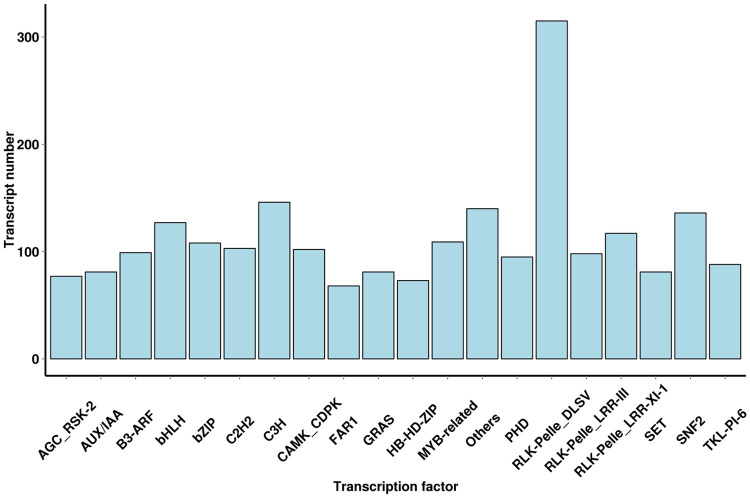
Transcription factors (TFs) play a very important role in the biological processes of plants, A total of 5,322 TFs were predicted by iTAK software, and the numbers of TFs enriched were as follows: RLK-pelle_DLSV (315), C3H (146), SNF2 (136), bHLH (127), and RLK-pelle_LRR-XI-1 (117) (Fig 6).
Fig 6. Transcript family of the distribution of TFs.
The x-axis represents the type of TF, and the y-axis represents the number of transcripts.
Discussion
The loquat genome has yet to be sequenced, research on the physiology and genetics mechanisms of this species has been restricted. Second generation sequencing technology is incapable of assembling full-length transcripts because of the shortness of sequencing reads. AS sites cannot be accurately detected, and the prediction accuracy is lower than 50% [41]. Moreover, fusion genes and gene families cannot be accurately detected. Thus, we can improve the accuracy of transcriptomic data and the prediction accuracy of AS by combining third-generation FL transcriptomic data with second-generation transcriptomic data. Third-generation combined with second-generation sequencing has been widely used to analyze rare transcripts, mining functional genes, analysing different genes in different tissues and at different developmental stages, and analysing the regulatory activity of TFs [42, 43]. To study plants for which a reference genome is not available, the most direct and effective use of ‘omics’ involves transcriptome and digital gene expression profile analysis [44], but obtaining high-quality reference genomes of genetically complex organisms remains costly and is technically challenging [45, 46]. In this study, a total of 13.41 Gb of raw data were obtained by SMRT sequencing, and after clustering analysis, non-FL sequence correction and the removal of redundant sequences, 38,435 transcripts with an average length of 2607 bp were obtained, which is far superior to previous studies of the loquat transcriptome using only the second-generation sequencing technique. For example,Song [47] obtained48,838 transcripts with an average length of 790 bp, and Xu [48] obtained 87,379 transcripts with an average length of 710 bp. Thus, Our findings indicated that SMRT sequencing is an effective route for obtaining reliable full-length transcript sequence information in plants.
LncRNAs are a class of non-coding RNA with a length longer than 200 nucleotides. Currently, many studies have been conducted to examine lncRNAs in animals [49–51], while research on lncRNAs in plants mainly focuses on a few model plants such as Arabidopsis thaliana [52], rice [53], and tomato [54]. In recent years, with the development of high-throughput sequencing technology, an increasing number of studies have focused on lncRNAs in plants, which have been found to play a regulatory role in plant flowering [55], reproductive development [56], photomorphogenesis [57], response to biotic and abiotic stresses [58], and in other biological processes [59]. In the present study, 407 lncRNAs were predicted from the non-redundant transcripts. These newly identifed lncRNAs will be helpful for loquat research in several aspects, and the function of lncRNAs in response to low temperature stress of loquat requires further study.
Full-length sequence transcripts are crucial for genome annotation and gene function research [60]. However, most methods for obtaining full-length transcripts are expensive, time-consuming and inefficient [61]. To date, no full-length sequence transcripts in loquat have been reported. In this study, 38,435 transcripts were obtained using the PacBio SMRT sequencing platform. Based on these transcripts, 37,230 ORFs were predicted, of which 27,905 had a complete CDS, and 37,993 transcripts were annotated into 8 databases including NR, eggNOG, Swiss-Prot, GO, COG, KOG, Pfam and KEGG. 37,908 transcripts annotated to the NR database, 46.22% of the sequences were aligned to Pyrus x and 45.40% to Malus domestica, whereas loquat itself had a best match percentage of 0.35%. These results may be due to the lack of transcript data related to loquat in the current NR database, reflecting the urgent need to improve the genetic database for this genus. The rational classification of protein coding is critical to maximize the use of transcripts for functional research. The results of the COG analysis showed that the R function (general function prediction only) constituted the greatest proportion, followed by the K function (transcription), L function (replication, recombination and repair) and T function (signal transduction mechanisms),which was similar to the results reported by Gong [17]. This result indicated that the gene expression of loquat under low-temperature stress is related to the above functions and suggested that the use of transcriptome sequencing technology is an effective method for the study of functional genes.
The results of this study provide a new reference for loquat transcription. However, analysis of the loquat transcriptome was not comprehensive, and gene expression and metabolic pathways associated with the mechanism underlying the cold stress response of loquat require further analysis.
Conclusion
This is the first study to perform SMRT sequencing of the FL transcriptome of embryos of young loquat fruit of plants under low-temperature stress. A total of 38,435 transcripts were obtained, 407 lncRNAs were predicted, 24,832 SSRs and 27,905 coding sequences were identified, and 37,993 transcripts were annotated for subsequent analysis. The number and average length of the transcripts were much better than those of previous studies in the loquat transcriptome using only the second-generation sequencing technique. SMRT sequencing is a useful and effective tool for acquiring reliable full-length transcripts of loquat. This work will facilitate research on the functional identification of genes and elucidation of the molecular mechanism underlying the cold stress response in loquat.
Acknowledgments
We would like to thank Biomarker Technologies Co., Ltd., for technical assistance with the RNA-Seq analysis. Professor Kevin is gratefully acknowledged for critical comments on the manuscript.
Data Availability
All the raw data was deposited into NCBI Sequence Read Archive(SRA) with accession number PRJNA623262 available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA623262.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Yang Q, Fu Y, Wang YQ, Deng QX, Tao L, Yan J, et al. Effects of simulated rain on pollen–stigma adhesion and fertilisation in loquat (Eriobotrya japonica Lindl.). J Hortic Sci Biotechnol. 2011; 86: 221–224. 10.1080/14620316.2011.11512751 [DOI] [Google Scholar]
- 2.Hong M, Chi ZH, Wang YQ, Tang YM, Deng QX, He MY, et al. Expression of a Chromoplast-Specific Lycopene beta-Cyclase Gene (CYC-B) Is Implicated in Carotenoid Accumulation and Coloration in the Loquat. Biomolecules. 2019; 9 10.3390/biom9120874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu H, Yang Y, Xie L, Li X, Feng C, Chen J, et al. Involvement of multiple types of dehydrins in the freezing response in loquat (Eriobotrya japonica). PLoS One. 2014; 9: e87575 10.1371/journal.pone.0087575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao S, Yang Z, Zheng Y. Sugar metabolism in relation to chilling tolerance of loquat fruit. Food Chem. 2013; 136: 139–143. 10.1016/j.foodchem.2012.07.113 [DOI] [PubMed] [Google Scholar]
- 5.Song H, Wang X, Hu W, Yang X, Diao E, Shen T, et al. A cold-induced phytosulfokine peptide is related to the improvement of loquat fruit chilling tolerance. Food Chem. 2017; 232: 434–442. 10.1016/j.foodchem.2017.04.045 [DOI] [PubMed] [Google Scholar]
- 6.Su W, Zhu Y, Zhang L, Yang X, Gao Y, Lin S. The cellular physiology of loquat (Eriobotrya japonica Lindl.) fruit with a focus on how cell division and cell expansion processes contribute to pome morphogenesis. Scientia Horticulturae. 2017; 224: 142–149. 10.1016/j.scienta.2017.06.012 [DOI] [Google Scholar]
- 7.Li G, Zhang Z, Yang X, Qiao Y, He X, Gao Y, et al. Inter-specific and Inter-generic Hybridization Compatibility of Eriobotrya Species (Loquat) and Related Genera. Hortic Plant J. 2016; 6: 315–322. 10.1016/j.hpj.2017.02.001 [DOI] [Google Scholar]
- 8.Wang L, Shao S, Madebo MP, Hou Y, Zheng Y, Jin P. Effect of nano-SiO2 packing on postharvest quality and antioxidant capacity of loquat fruit under ambient temperature storage. Food Chem. 2020; 315: 126295 10.1016/j.foodchem.2020.126295 [DOI] [PubMed] [Google Scholar]
- 9.Papadakis IE, Tsiantas PI, Tsaniklidis G, Landi M, Psychoyou M, Fasseas C. Changes in sugar metabolism associated to stem bark thickening partially assist young tissues of Eriobotrya japonica seedlings under boron stress. J Plant Physiol. 2018; 231: 337–345. 10.1016/j.jplph.2018.10.012 [DOI] [PubMed] [Google Scholar]
- 10.Liu C, Wang M, Wang L, Guo Q, Liang G. Extensive genetic and DNA methylation variation contribute to heterosis in triploid loquat hybrids. Genome. 2018; 61: 437–447. 10.1139/gen-2017-0232 [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Liu T, Ohlson EW, Wang L, Wu D, Guo Q, et al. Loquat (Eriobotrya japonica (Thunb.) circadian clock gene cloning and heterosis studies of artificial triploid loquat. Sci Hortic. 2019; 246: 328–337. 10.1016/j.scienta.2018.10.068 [DOI] [Google Scholar]
- 12.Sanhong W, Qian W, Ying Z, Hongli Q, Huakun W. Identification of two new S-RNases and molecular S-genotyping of twenty loquat cutivars [Eriobotrya japonica (Thunb.) Lindl.]. Sci Hortic. 2017; 218: 48–55. 10.1016/j.scienta.2017.02.002 [DOI] [Google Scholar]
- 13.Jing D, Chen W, Xia Y, Shi M, Wang P, Wang S, et al. Homeotic transformation from stamen to petal in Eriobotrya japonica is associated with hormone signal transduction and reduction of the transcriptional activity of EjAG. Physiol Plant. 2020; 168: 893–908. 10.1111/ppl.13029 [DOI] [PubMed] [Google Scholar]
- 14.Hadjipieri M, Georgiadou EC, Marin A, Diaz-Mula HM, Goulas V, Fotopoulos V, et al. Metabolic and transcriptional elucidation of the carotenoid biosynthesis pathway in peel and flesh tissue of loquat fruit during on-tree development. BMC Plant Biol. 2017; 17: 102 10.1186/s12870-017-1041-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang S, Luo J, Xu F, Zhang X. Transcriptome Analysis Reveals Candidate Genes Involved in Gibberellin-Induced Fruit Setting in Triploid Loquat (Eriobotrya japonica). Front Plant Sci. 2016; 7: 1924 10.3389/fpls.2016.01924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin S, Wu T, Lin H, Zhang Y, Xu S, Wang J, et al. De Novo Analysis Reveals Transcriptomic Responses in Eriobotrya japonica Fruits during Postharvest Cold Storage. Genes (Basel). 2018; 9 10.3390/genes9120639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong RG, Lai J, Yang W, Liao MA, Wang ZH, Liang GL. Analysis of alterations to the transcriptome of Loquat (Eriobotrya japonica Lindl.) under low temperature stress via de novo sequencing. Genet Mol Res. 2015; 14: 9423–9436. 10.4238/2015.August.14.6 [DOI] [PubMed] [Google Scholar]
- 18.Zeng D, Chen X, Peng J, Yang C, Peng M, Zhu W, et al. Single-molecule long-read sequencing facilitates shrimp transcriptome research. Sci Rep. 2018; 8: 16920 10.1038/s41598-018-35066-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding N, Wang A, Zhang X, Wu Y, Wang R, Cui H, et al. Identification and analysis of glutathione S-transferase gene family in sweet potato reveal divergent GST-mediated networks in aboveground and underground tissues in response to abiotic stresses. BMC Plant Biol. 2017; 17: 225 10.1186/s12870-017-1179-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao Q, Gao ZF, Zhang D, Zhao BG, Dong FQ, Fu CX, et al. The developmental dynamics of the Populus stem transcriptome. Plant Biotechnol J. 2018; 17: 206–219. 10.1111/pbi.12958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdel-Ghany SE, Hamilton M, Jacobi JL, Ngam P, Devitt N, Schilkey F, et al. A survey of the sorghum transcriptome using single-molecule long reads. Nat Commun. 2016; 7: 11706 10.1038/ncomms11706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B, Tseng E, Regulski M, Clark TA, Hon T, Jiao Y, et al. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat Commun. 2016; 7: 11708 10.1038/ncomms11708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M, Wang P, Liang F, Ye Z, Li J, Shen C, et al. A global survey of alternative splicing in allopolyploid cotton: landscape, complexity and regulation. New Phytol. 2018; 217: 163–178. 10.1111/nph.14762 [DOI] [PubMed] [Google Scholar]
- 24.Shin SC, Ahn DH, Kim SJ, Lee H, Oh TJ, Lee JE, et al. Advantages of Single-Molecule Real-Time Sequencing in High-GC Content Genomes. PLoS One. 2013; 8: e68824 10.1371/journal.pone.0068824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009; 323: 133–138. 10.1126/science.1162986 [DOI] [PubMed] [Google Scholar]
- 26.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006; 22: 1658–1659. 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- 27.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015; 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25: 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Mei W, Soltis PS, Soltis DE, Barbazuk WB. Detecting alternatively spliced transcript isoforms from single-molecule long-read sequences without a reference genome. Mol Ecol Resour. 2017; 17: 1243–1256. 10.1111/1755-0998.12670 [DOI] [PubMed] [Google Scholar]
- 30.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013; 8: 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007; 35: W345–349. 10.1093/nar/gkm391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013; 41: e74 10.1093/nar/gkt006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng Y, Li JQ, Wu SF, Zhu YP, Chen YW, He FC, et al. Integrated NR Database in Protein Annotation System and Its Localization. Computer Engineering. 2006; 32: 71–74. [Google Scholar]
- 34.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004; 32: D115–119. 10.1093/nar/gkh131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000; 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000; 28: 33–36. 10.1093/nar/28.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004; 5: R7 10.1186/gb-2004-5-2-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44: D279–285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004; 32: D277–280. 10.1093/nar/gkh063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng Y, Jiao C, Sun H, Rosli HG, Pombo MA, Zhang P, et al. iTAK: A Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol Plant. 2016; 9: 1667–1670. 10.1016/j.molp.2016.09.014 [DOI] [PubMed] [Google Scholar]
- 41.Wang T, Wang H, Cai D, Gao Y, Zhang H, Wang Y, et al. Comprehensive profiling of rhizome-associated alternative splicing and alternative polyadenylation in moso bamboo (Phyllostachys edulis). Plant J. 2017; 91: 684–699. 10.1111/tpj.13597 [DOI] [PubMed] [Google Scholar]
- 42.Liu W, Xiong C, Yan L, Zhang Z, Ma L, Wang Y, et al. Transcriptome Analyses Reveal Candidate Genes Potentially Involved in Al Stress Response in Alfalfa. Front Plant Sci. 2017; 8: 26 10.3389/fpls.2017.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Workman RE, Myrka AM, Wong GW, Tseng E, Welch KC Jr., Timp W. Single-molecule, full-length transcript sequencing provides insight into the extreme metabolism of the ruby-throated hummingbird Archilochus colubris. Gigascience. 2018; 7: 1–12. 10.1093/gigascience/giy009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009; 10: 57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dufresne F, Stift M, Vergilino R, Mable BK. Recent progress and challenges in population genetics of polyploid organisms: an overview of current state-of-the-art molecular and statistical tools. Mol Ecol. 2014; 23: 40–69. 10.1111/mec.12581 [DOI] [PubMed] [Google Scholar]
- 46.Spannagl M, Martis MM, Pfeifer M, Nussbaumer T, Mayer KF. Analysing complex Triticeae genomes—concepts and strategies. Plant Methods. 2013; 9: 35 10.1186/1746-4811-9-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song H, Zhao X, Hu W, Wang X, Shen T, Yang L. Comparative Transcriptional Analysis of Loquat Fruit Identifies Major Signal Networks Involved in Fruit Development and Ripening Process. Int J Mol Sci. 2016; 17 10.3390/ijms17111837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu HX, Li XY, Chen JW. Comparative transcriptome profiling of freezing stress responses in loquat (Eriobotrya japonica) fruitlets. J Plant Res. 2017; 130: 893–907. 10.1007/s10265-017-0942-4 [DOI] [PubMed] [Google Scholar]
- 49.Li K, Tian Y, Yuan Y, Fan X, Yang M, He Z, et al. Insights into the Functions of LncRNAs in Drosophila. Int J Mol Sci. 2019; 20 10.3390/ijms20184646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pegueroles C, Iraola-Guzman S, Chorostecki U, Ksiezopolska E, Saus E, Gabaldon T. Transcriptomic analyses reveal groups of co-expressed, syntenic lncRNAs in four species of the genus Caenorhabditis. RNA Biol. 2019; 16: 320–329. 10.1080/15476286.2019.1572438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bush SJ, Muriuki C, McCulloch MEB, Farquhar IL, Clark EL, Hume DA. Cross-species inference of long non-coding RNAs greatly expands the ruminant transcriptome. Genet Sel Evol. 2018; 50: 20 10.1186/s12711-018-0391-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Severing E, Faino L, Jamge S, Busscher M, Kuijer-Zhang Y, Bellinazzo F, et al. Arabidopsis thaliana ambient temperature responsive lncRNAs. BMC Plant Biol. 2018; 18: 145 10.1186/s12870-018-1362-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H, Wang R, Mao B, Zhao B, Wang J. Identification of lncRNAs involved in rice ovule development and female gametophyte abortion by genome-wide screening and functional analysis. BMC Genomics. 2019; 20: 90 10.1186/s12864-019-5442-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cui J, Jiang N, Hou X, Wu S, Zhang Q, Meng J, et al. Genome-Wide Identification of lncRNAs and Analysis of ceRNA Networks During Tomato Resistance to Phytophthora infestans. Phytopathology. 2020; 110: 456–464. 10.1094/PHYTO-04-19-0137-R [DOI] [PubMed] [Google Scholar]
- 55.Zhao X, Li J, Lian B, Gu H, Li Y, Qi Y. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat Commun. 2018; 9: 5056 10.1038/s41467-018-07500-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Fan X, Lin F, He G, Terzaghi W, Zhu D, et al. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci U S A. 2014; 111: 10359–10364. 10.1073/pnas.1409457111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, et al. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci U S A. 2012; 109: 2654–2659. 10.1073/pnas.1121374109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui J, Luan Y, Jiang N, Bao H, Meng J. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 2017; 89: 577–589. 10.1111/tpj.13408 [DOI] [PubMed] [Google Scholar]
- 59.Wu HW, Deng S, Xu H, Mao HZ, Liu J, Niu QW, et al. A noncoding RNA transcribed from the AGAMOUS (AG) second intron binds to CURLY LEAF and represses AG expression in leaves. New Phytol. 2018; 219: 1480–1491. 10.1111/nph.15231 [DOI] [PubMed] [Google Scholar]
- 60.Rhoads A, Au KF. PacBio Sequencing and Its Applications. Genomics Proteomics Bioinformatics. 2015; 13: 278–289. 10.1016/j.gpb.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bayega A, Wang YC, Oikonomopoulos S, Djambazian H, Fahiminiya S, Ragoussis J. Transcript Profiling Using Long-Read Sequencing Technologies. Methods Mol Biol. 2018; 1783: 121–147. 10.1007/978-1-4939-7834-2_6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the raw data was deposited into NCBI Sequence Read Archive(SRA) with accession number PRJNA623262 available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA623262.