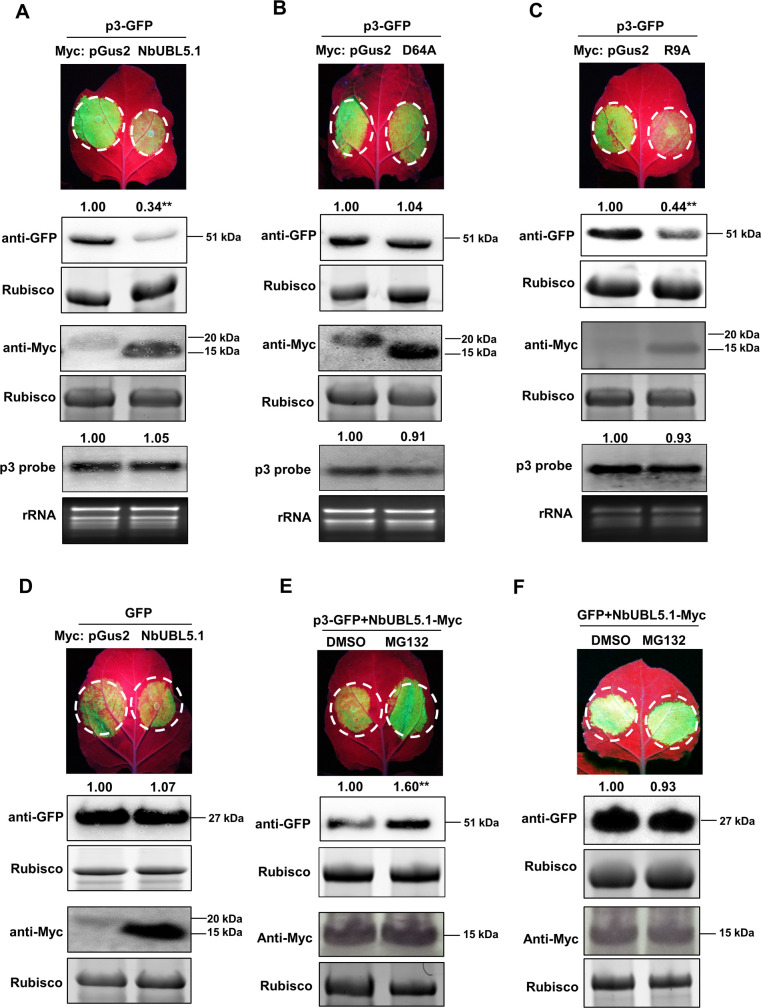
Fig 4. NbUBL5.1 mediates p3 degradation through the 26S proteasome system.
A. GFP-fused p3 was co-expressed with Myc-fused NbUBL5.1 or Myc-fused pGus2 as control in leaves of wild type N. benthamiana through agroinfiltration. At 3.5 dpi, fluorescence and accumulation of GFP-fused p3 protein in zones co-expressing NbUBL5.1-Myc was notably reduced, compared to that in zones expressing pGus2-Myc with p3-GFP. B and C. Effect of NbUBL5.1 mutants, D64A(B) and R9A(C) on p3-GFP accumulation. Fluorescence and accumulation of GFP-fused p3 protein in zones co-expressing R9A-Myc was notably reduced, while those in zones expressing D64A-Myc with p3-GFP were at similar levels to the control. D. Myc-fused NbUBL5.1 did not affect fluorescence and protein accumulation of GFP in the control experiments. E. NbUBL5.1-Myc and p3-GFP were co-expressed in leaves treated with MG132 or DMSO. At 4 dpi, fluorescence and accumulation of GFP-fused p3 protein in zones treated with MG132 were greater than those in zones treated with DMSO. F. MG132 treatment had no effect on the fluorescence and protein accumulation of GFP in the presence of NbUBL5.1-Myc. Expression of the Myc-fused proteins was confirmed by western blot using a Myc antibody. Results are from three biological replicates. Band intensity in blots was calculated by ImageJ based on three replicates. A two-sample unequal variance directional t test was used to test the significance of the difference (**, p value<0.01).