Figure 6.
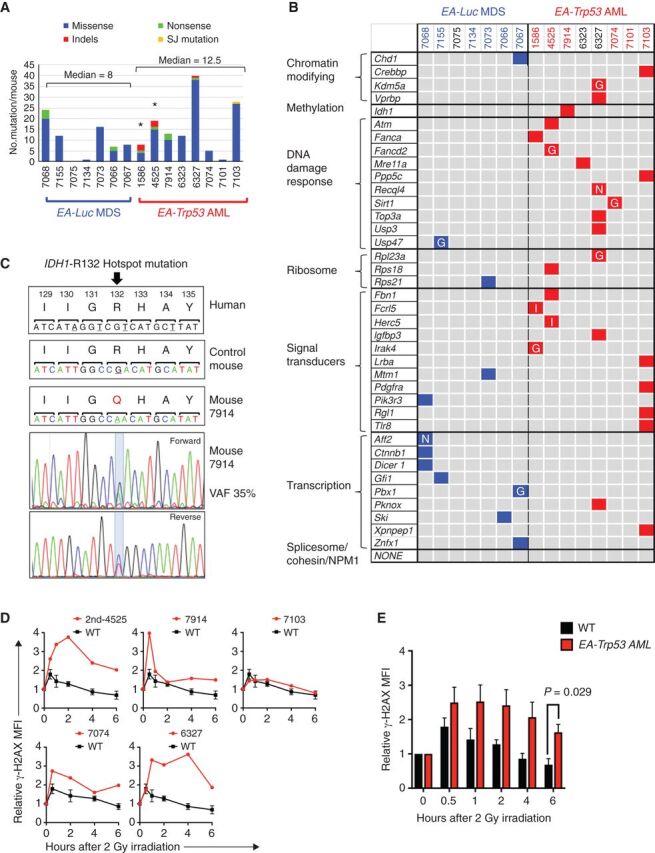
Enrichment of DNA repair gene mutations in myeloid neoplasms with Trp53 knockdown. A, Summary of nonsynonymous mutations in 7 EA-Luc MDS and 8 EA, Trp53-AML samples. Both donor and recipient mice were treated with ENU with the exception of 3 mice: 1586 and 4525 (no ENU); 7914 (ENU recipient alone). All EA, Trp53 mice had a >20% blast count, except 6327, which had 18% blasts. The median number of mutations was not statistically different in the EA-Luc MDS versus the EA-Trp53 AML group (8 vs. 12.5). Two EA, Trp53-AMLs (1586, 4525), which arose without ENU treatment (*) had a median of 13.5 mutations. B, Because there were no genes that were mutated in multiple mice, mutations were classified into 10 gene categories that are commonly mutated (somatic and inherited) in patients with MDS or AML. Colored boxes identify genes that are mutated in each mouse. Nonsense (N) and indel (I) mutations are indicated. Most missense mutations were considered to be pathogenic by at least two of the following algorithms: GERP, SIFT, and PolyPhenv2 (Supplementary Table S4). Missense mutations considered pathogenic by GERP score alone are indicated by the letter “G.” With the exception of 7101, all EA, Trp53-AMLs harbor a mutation in a gene within the gene ontology term “DNA repair.” Bone marrow samples from mice identified in blue and red numbering were used for RNA-seq analysis. C, The normal nucleotide and amino acid sequence and position (human) of the IDH1 gene are shown in the upper panels. The amino acid sequence is conserved in this region. Mouse 7914 had a mutation at the R132 position, which is a hotspot mutation in humans. The chromatograms with the mutation are shown. D, γH2AX levels were measured in CD117 (Kit)+ (4525-2nd, 7914) or CD11b+ (7074, 6327, 7103, 7098) cells at baseline, and up to 6 hours post 2 Gy irradiation (4525-2nd and 7914 expressed Kit, but not CD11b). We previously showed that leukemia cells from 1586 and 4525 have an aberrant DNA damage response (6). AML that arose in a secondary transplant using 4525 donor cells (4525-2nd sample) also showed a defective DNA repair response. The mean fluorescence intensity measured using flow cytometry, was normalized to the 0-hour time point for each sample, to compare the induction and resolution kinetics among samples. Kinetics of each AML sample is shown in comparison to the average kinetics of CD11b+ wild-type cells. We could not assess the DNA damage response in 7101 (no DDR mutation), because the irradiation did not induce a γH2AX signal at any time point. E, The mean γH2AX levels for WT (n = 3) and EA, Trp53-AML (n = 5) samples ± SEM are shown. An independent, two-tailed t test was performed for each time point. The difference was statistically significant at the 6-hour time point.
