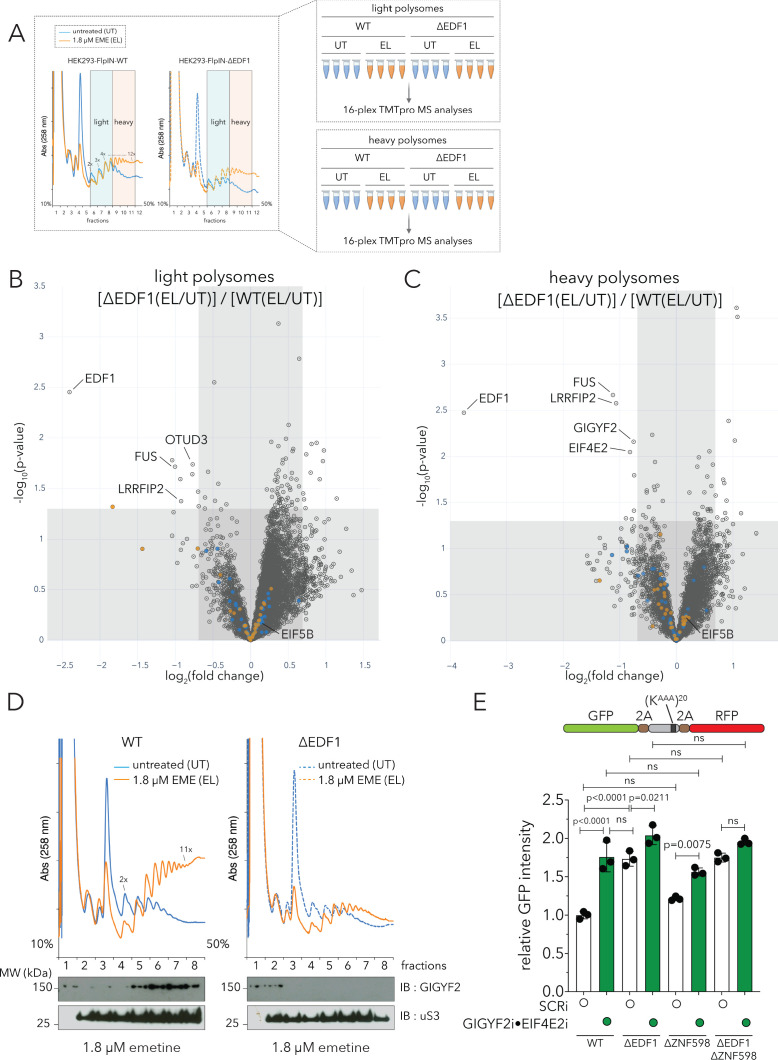
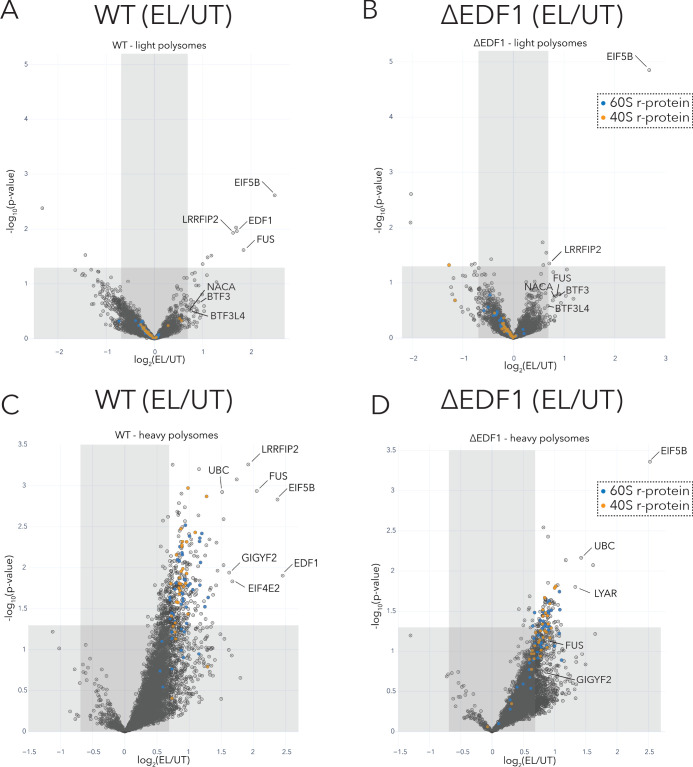
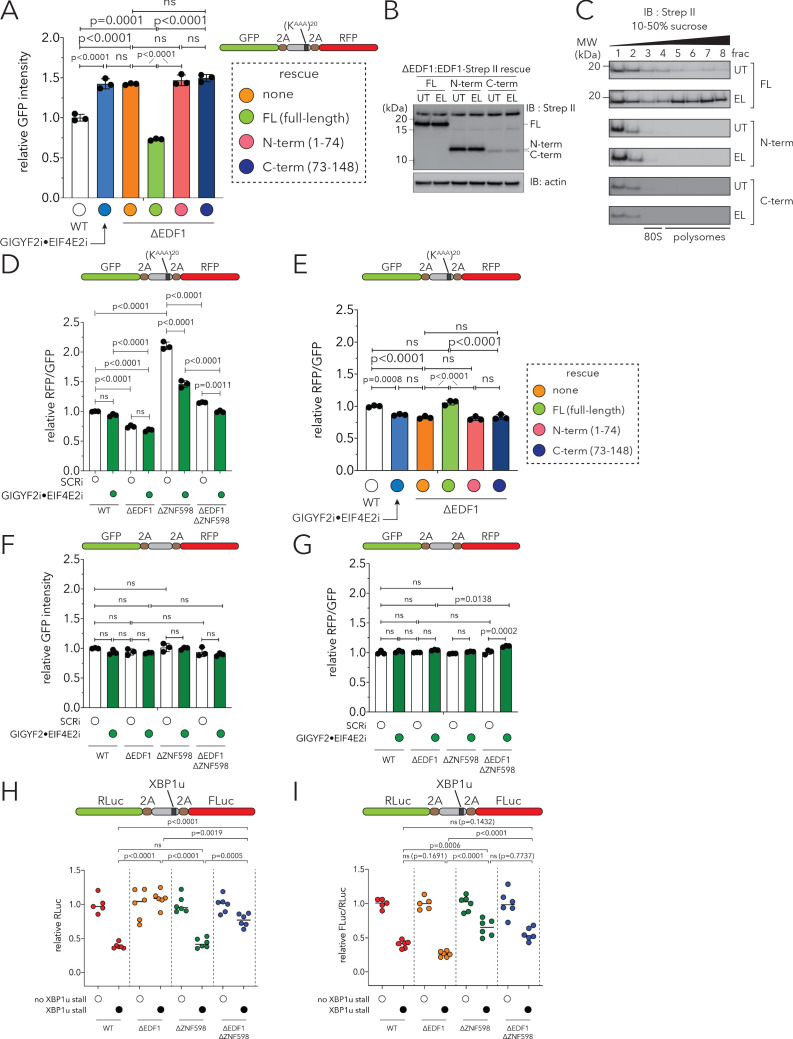
Figure 6. EDF1 recruits GIGYF2•EIF4E2 to collided ribosomes.
(A) (Left) UV (A258) absorbance across 10–50% sucrose gradients from lysates of HEK293 Flp-In TREx WT and ΔEDF1 cells left untreated (UT, blue trace) or treated with 1.8 µM emetine (EL, orange trace) for 15 min; fractions 6–8 pooled for light polysomes; fractions 9–11 pooled for heavy polysomes (n = 4). (Right) Schematic of polysome proteomics pipeline to monitor relative change in protein intensity in response to emetine treatment in light and heavy polysomes between WT and ΔEDF1 cells. (B, C) Volcano plot of log2 indicated ratio (x-axis) against -log10(p-value) (y-axis) for light (B) or heavy (B) polysomes. (D) UV (A258) absorbance across 10–50% sucrose gradients from lysates of HEK293-Flp-In TREx WT and ΔEDF1 cells left untreated (UT, blue trace) or treated with 1.8 µM emetine (EL, orange trace) for 15 min; TCA precipitated proteins from individual fractions were resolved by SDS-PAGE and analyzed by immunoblotting using GIGYF2 and uS3 antibodies. (n = 2) (E) Relative GFP intensity from HEK293 Flp-In TREx WT, ΔEDF1, ΔZNF598, and ΔEDF1ΔZNF598 cells transfected with the GFP-(KAAA)20-RFP stalling reporter without (white bars, non-targeting siRNA, SCRi) or with siRNA-mediated depletion of GIGYF2 and EIF4E2 (green bars; GIGYF2i•EIF4E2i). Error bars denote SD for n = 3. p-values were determined by one-way ANOVA and Tukey’s post hoc correction for multiple comparisons. See also Figure 6—figure supplement 1, Figure 6—figure supplement 2 and Figure 6—source data 1.