Figure 2.
CRISPR/Cas9-Mediated Excision of Whole Ctnnb1 Locus Results in a Complete β-Catenin Knockout Model in mESCs
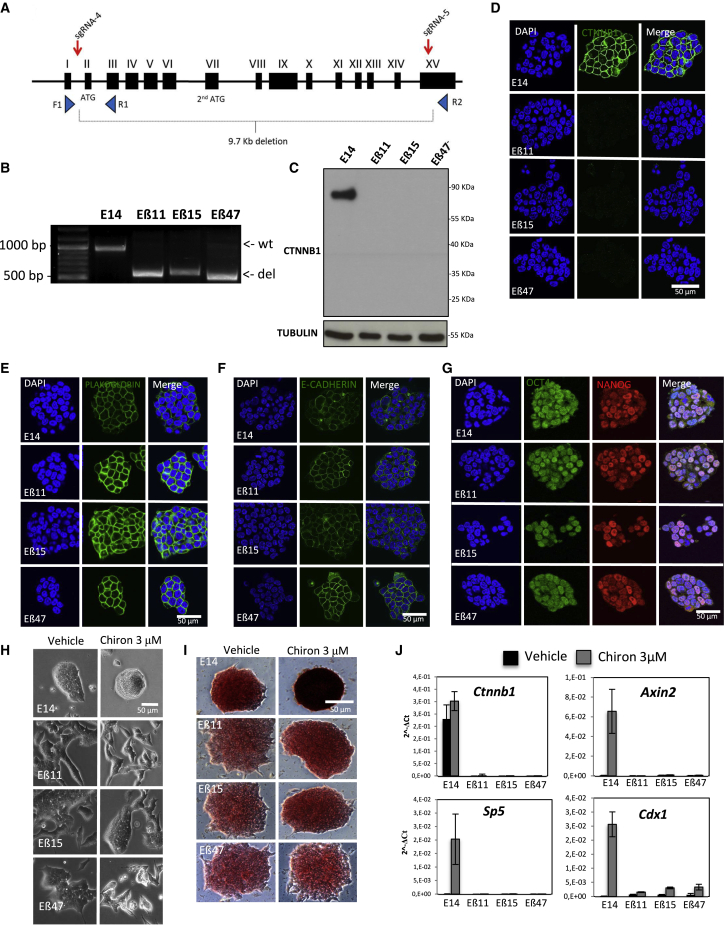
(A) Schematic representation of sgRNA design for CRISPR/Cas9-mediated excision of whole β-catenin coding sequence. Red arrows indicate sgRNAs target sites. Blue triangles indicate position and orientation of oligonucleotides used for PCR genotyping.
(B) PCR genotyping of three homozygous β-catenin knockout clones (Eβ11, Eβ15, and Eβ47) and parental E14 mESCs. Expected amplicon size is 951 bp for wild-type (wt) alleles and 551 bp for knockout alleles (del).
(C) Western blot of total protein extracts from Eβ11, Eβ15, Eβ47, and wild-type E14 cells. Protein extracts were probed for β-catenin (using a C-terminally raised antibody), stripped and re-probed for TUBULIN as loading control.
(D–G) Immunofluorescence in fixed parental E14, Eβ11, Eβ15, and Eβ47 cells for β-catenin (D), PLAKOGLOBIN (E), E-CADHERIN (F), OCT4 and NANOG (G). DAPI was used to counterstain nuclei. Scale bar represents 50 μm.
(H) Phase contrast pictures of Eβ11, Eβ15, Eβ47, and parental E14 clones upon 72-h vehicle (0.3% DMSO, left) or Chiron 3 μM treatment (right) in serum/LIF. Cells were seeded at 4 × 105 cells/well density in 6-well plates. Scale bar represents 50 μm.
(I) Phase contrast pictures of AP staining on E14, Eβ11, Eβ15, and Eβ47 cells cultured in serum/LIF in presence of vehicle (0.3% DMSO, left) or Chiron 3 μM (right) for 5 days; 300 cells were seeded in each well of a 6-well plate. Scale bar represents 50 μm.
(J) Histogram of qRT-PCR data on total RNA extracts of E14, Eβ11, Eβ15, and Eβ47 cells exposed to vehicle (0.3% DMSO, black bars) or Chiron 3 μM; 2−ΔCt are represented, and GAPDH was used as internal control. Error bars represents standard deviations of three technical replicates.