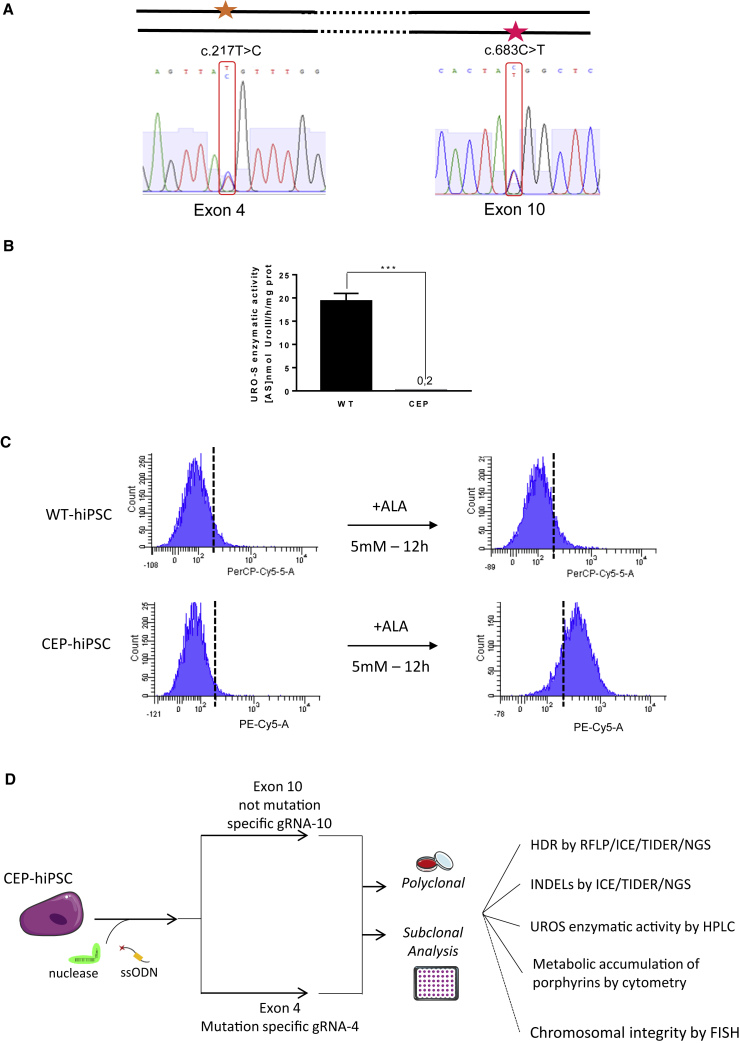
Figure 1.
CEP-hiPSC Characterization
(A) CEP-hiPSC UROS genotyping. The CEP-hiPSC is composite heterozygous, with one allele mutated in exon 4 (c.217T > C) and the other one on the exon 10 (c.683C > T).
(B) WT-hiPSC and CEP-hiPSC UROS enzymatic activity. Quantification of UROS enzymatic activity by high-performance liquid chromatography (HPLC) of WT or CEP-hiPSC. Data are represented as mean ± SEM. n ≥ 5 independent experiments for each IPSC lines.
(C) WT-hiPSC and CEP-hiPSC metabolic activity. Type I fluorocyte accumulation (PE-Cy5-A) by flow cytometry: illustrative results of WT (top) and CEP-hiPSC (bottom) before and after δ-aminolevulinic acid (ALA) addition (5 mM, 12 h).
(D) Experimental workflow for UROS gene editing and result analysis. CEP-hiPSCs were transfected by nucleofection with a fluorescent ssODN template, a ZsGreen-nuclease plasmid, and an RNA guide plasmid targeting exon 4 or 10. Then polyclonal and further monoclonal analysis were performed: HDR was quantified by RFLP, and confirmed by ICE and NGS. The indels were quantified by ICE and NGS. The UROS functionality was assessed by quantifying UROS-specific activity by HPLC and type I porphyrin accumulation by flow cytometry. For the subclones, chromosomal integrity was checked by FISH (analysis only performed for subclones).