Figure 2.
Polyclonal Analysis of UROS Exon 10 Editing by CRISPR/Cas9
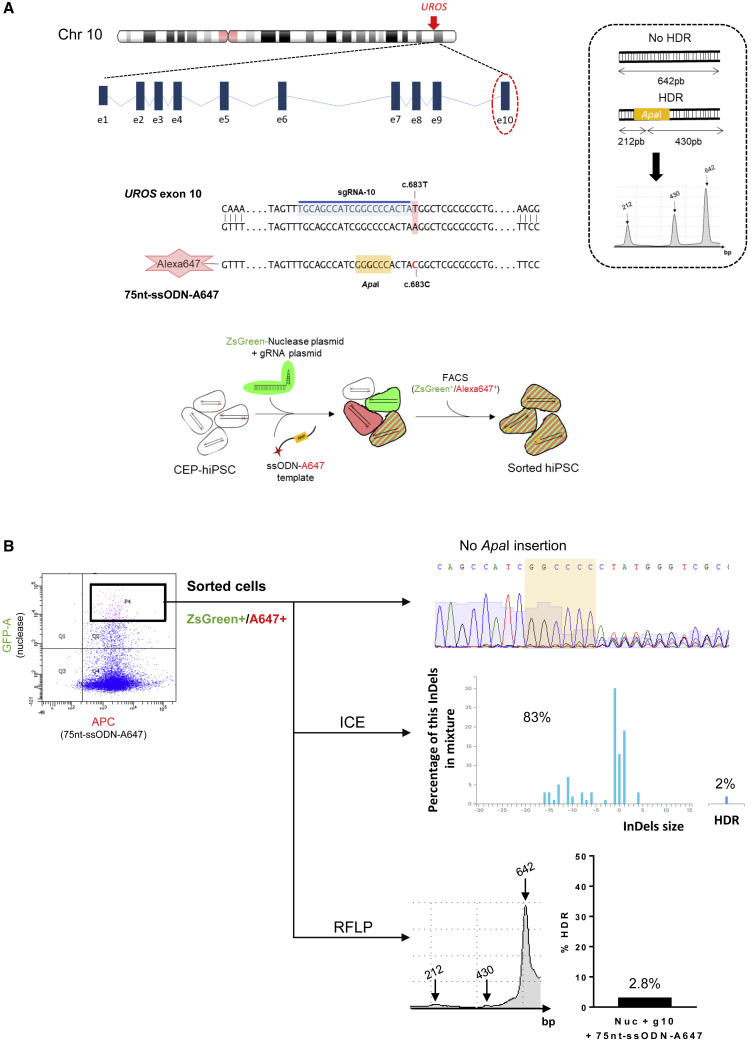
(A) Top, schematic UROS locus on chromosome 10 with UROS gene overview and the exon 10 targeted region. Middle, detailed view of exon 10 region close to the c.683T mutation. CRISPR-mediated HDR design using an sgRNA targeting the sequence just next to the mutation (sgRNA-10 highlighted in blue) and a 75nt-ssODN-A647 carrying a silent restriction site ApaI (highlight in yellow) and the correction of the mutation (c.683C in red). Right, scheme of ApaI-digested PCR products obtained for alleles with or without HDR and an illustrative RFLP analysis. Bottom, exon 10 gene editing workflow. The CEP-hiPSCs were transfected with a ZsGreen-nuclease plasmid, the sgRNA-10 plasmid, and a 75nt-ssODN-A647 template. The cells were sorted by cytometry to get back only the cells transfected with the ZsGreen-nuclease and the 75nt-ssODN-A647 template.
(B) Flow cytometry illustration of the double-positive ZsGreen+/A647+ sorted cells (left) and their targeting region analysis: Sanger sequence (top), ICE (middle), and RFLP (bottom).