Extended data Fig. 9 |. Additional nucleolar organization and sequencing analyses related to Ewing sarcoma.
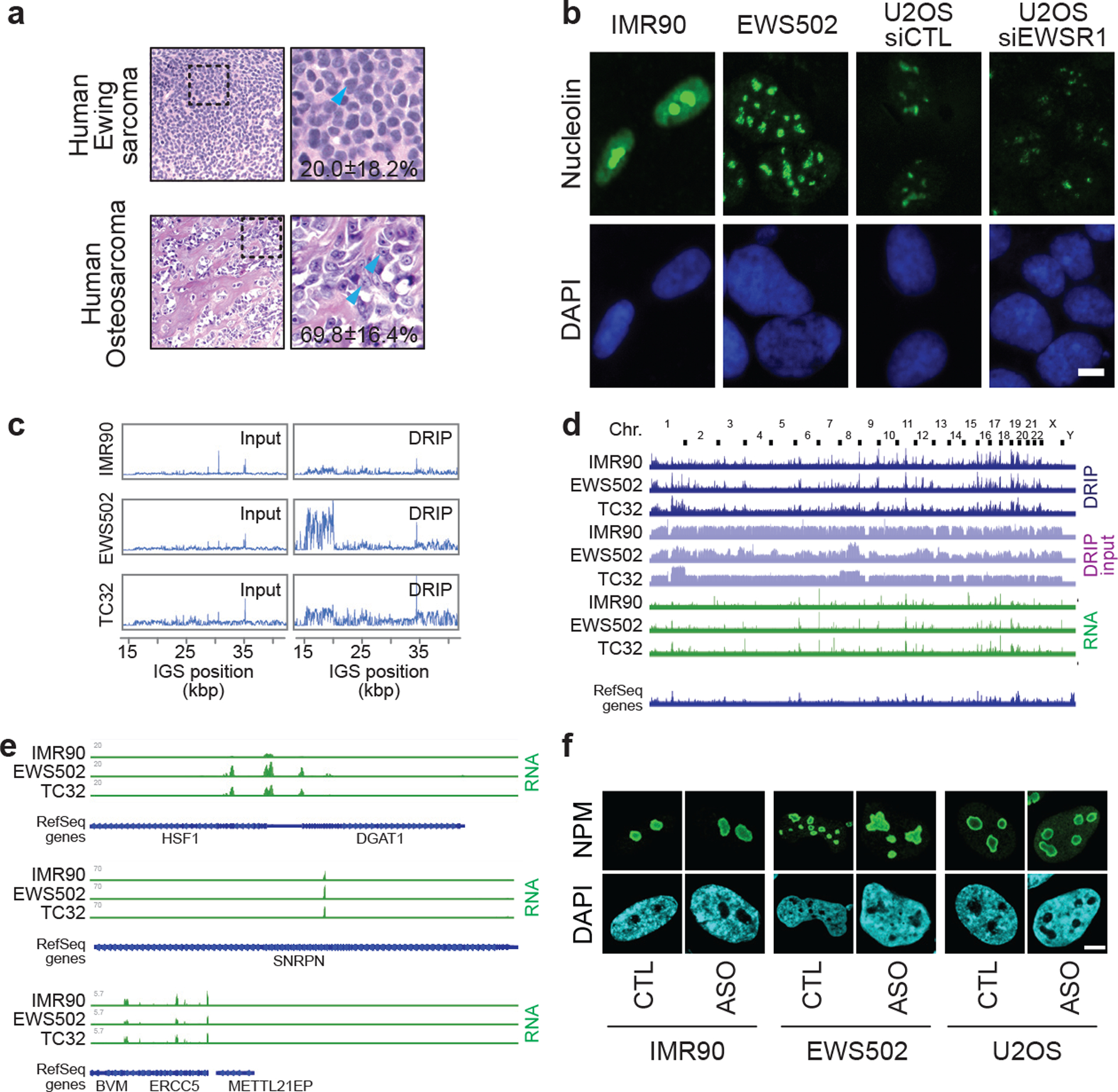
(a) Representative tissue sections of human Ewing sarcoma and osteosarcoma (haematoxylin and eosin, x400). Materials were obtained following Institutional Research Ethics Board approval (Sinai Heath Systems, 17–0103-E). The percentage of cells with one or two distinct nucleoli per nucleus is shown. Data are shown as the mean±s.d.; per cancer type, n=5 cases (100 cells each); two-tailed t-test P=0.0019. (b) Ewing sarcoma cells (EWS502), and U2OS cells with siEWSR1 display disrupted nucleoli, as indicated by the nucleolin protein, compared to their respective control IMR90 and U2OS siControl (siCTL) cells. Scale bar, 5 μm. (c) Ewing sarcoma (EWS502, TC32) cells showed increased R-loop levels across IGSs in DRIP-seq. (d) Genome-wide view of sequence read alignments for DRIP-seq and RNA-seq. Chr., chromosome. (e) IMR90, EWS502, and TC32 cells can exhibit similarities and differences at non-rDNA loci in sequencing read alignments from RNA-seq. (f) ASO targeting sincRNAs ameliorates nucleolar organization. Shown are representative images related to quantifications in Fig. 4d. Images are representative of two independent experiments. Scale bar, 5 μm.
