Fig. 4. Nucleolar Pol II reinforcement by SETX and nucleolus-disrupting sincRNAs in cancer.
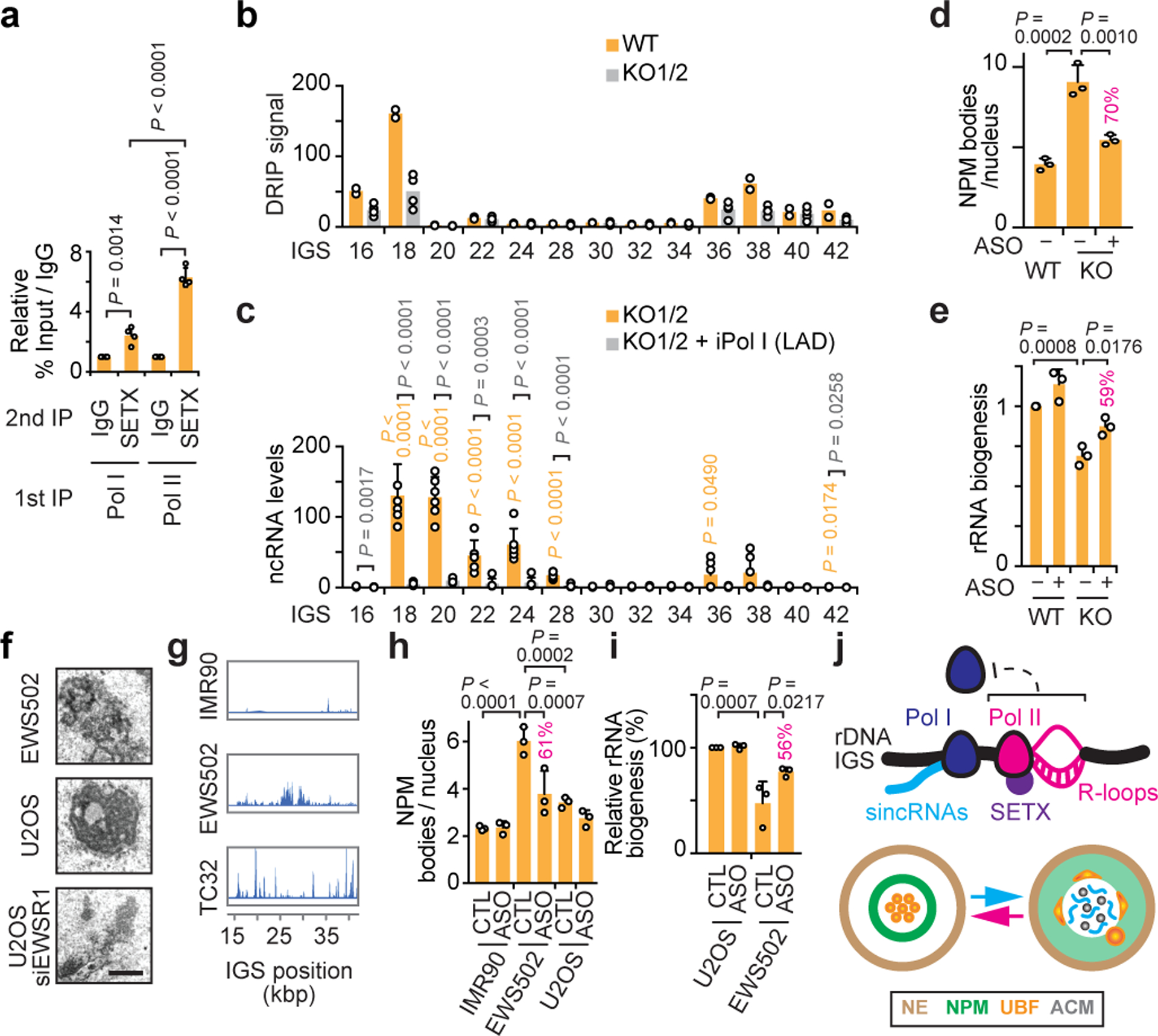
(a) Sequential immunoprecipitations (IPs) revealed preferential SETX co-enrichment with Pol II at IGSs. (b-c) SETX KO in two clones decreased R-loops (b) and induced IGS ncRNAs (c). (d-e) In SETX KO cells, single cell analysis showed that ASO-mediated repression of Pol I-dependent sincRNAs partly rescues nucleolar organization (d) and pre-rRNA processing (e). Percentages indicating the magnitude of ASO-mediated phenotypic rescue are shown above graph bars where applicable. (f-g) Ewing sarcoma (EWS502, TC32) cells showed both disrupted nucleoli by electron microscopy (f) and increased ncRNA levels across IGSs (g). (h,i) Single-cell analysis showed that sincRNA knockdown partly restores nucleolar organization (h) and pre-rRNA processing (i) in EWS502 cells. (j) Model showing Pol II-dependent R-loop shield limiting Pol I-dependent sincRNAs, which compromise nucleolar organization and function. NE, nuclear envelope; ACM, amyloid-converting motif. (a-i) HEK293 cells; data are shown as the mean±s.d.; one-way ANOVA with Dunnett’s multiple comparisons test (a, d-e, h-i) and one-way ANOVA with Tukey’s multiple comparisons test (c); n=4 biologically independent experiments (a), n=2 biologically independent experiments (b, duplicates for each of WT, KO1, and KO2), n=6 biologically independent experiments (c, triplicates for each KO), n=3 biologically independent experiments (d-e,h-i); images in (f-g) are representatives of two independent experiments, scale bar, 1 μm.
