Figure 3.
Comparison of the HA-Binding Modes among Different VH6-1-Encoded bnAbs
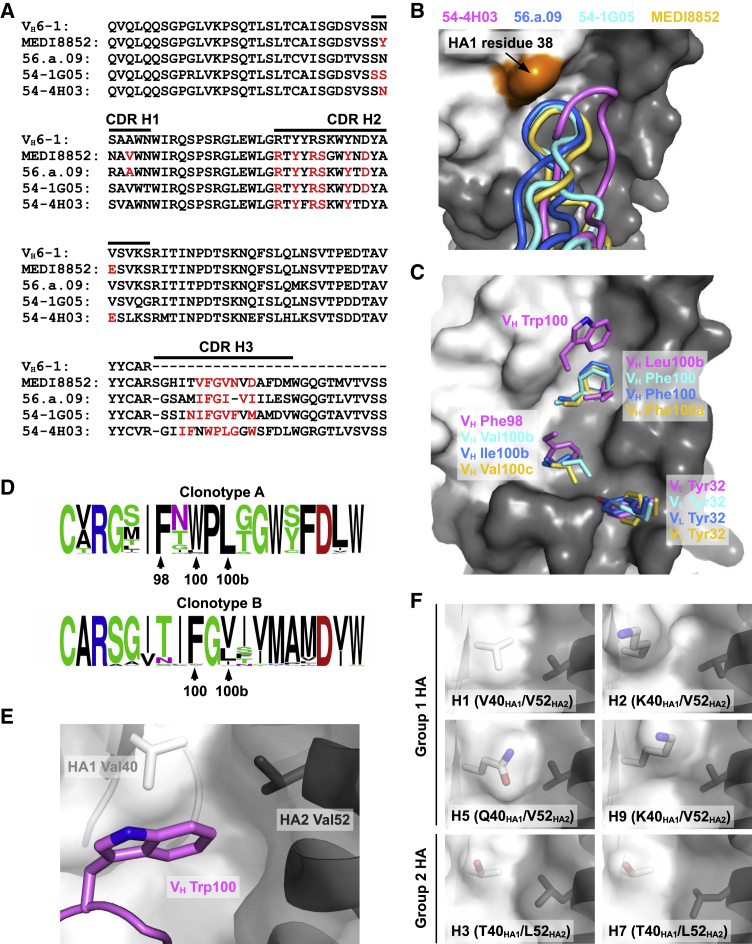
(A) Alignment of the germline VH6-1 sequence with those of 54-4H03, 54-1G05, 56.a.09 (Joyce et al., 2016), and MEDI8852 (Kallewaard et al., 2016). The regions that correspond to CDR H1, H2, and H3 (Kabat numbering scheme) are indicated. Paratope residues are highlighted in red.
(B) Conformations of CDR H3 from 54-4H03, 54-1G05, 56.a.09 (PDB 5K9K) (Joyce et al., 2016), and MEDI8852 (PDB 5JW4) (Kallewaard et al., 2016) are compared. HA1 surface is colored in white, HA2 in gray, and HA1 residue 38 in orange.
(C) Residues on CDR H3 of 54-4H03, 54-1G05, MEDI8852 (PDB 5JW4) (Kallewaard et al., 2016), and 56.a.09 (PDB 5K9K) (Joyce et al., 2016) that are important for binding to the HA stem are shown. VL Tyr32 on CDR L1 that is also important for binding is shown. The HA1 surface is colored in white and HA2 in gray.
(D) Sequence variation of CDR H3 among members of clonotype A (including 54-4H03) and clonotype (including 54-1G05) are shown as sequence logos. The relative sizes of the letters represent their occurrence frequency.
(E) Interaction between VH Trp100 on CDR H3 of 54-4H03 with HA of A/California/04/2009 (H1N1) is highlighted.
(F) Shapes of the binding pocket for accommodating VH Trp100 of 54-4H03 among different HA subtypes are compared. H1 HA: PDB 3LZG (Xu et al., 2010a). H2 HA: PDB 3KU5 (Xu et al., 2010b). H3 HA: PDB 4FNK (Ekiert et al., 2012). H5 HA: PDB 4BGW (Xiong et al., 2013). H7 HA: PDB 4LN6 (Yang et al., 2013). H9 HA: PDB 1JSD (Ha et al., 2002).