Figure 3.
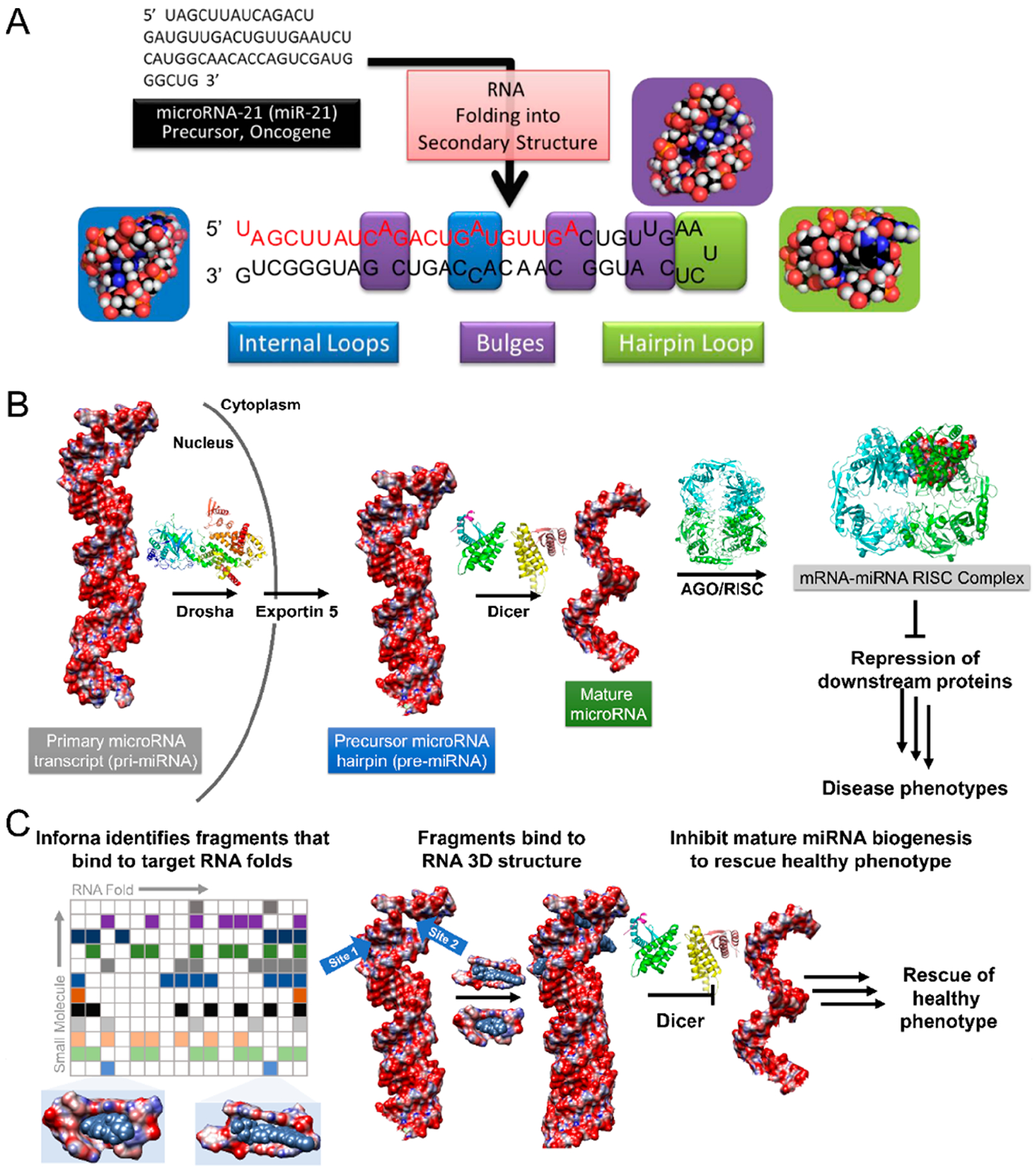
Modular RNA secondary structure motifs form three-dimensional structures. (A) Hierarchical assembly of RNA structure from sequence to secondary structure. Many of these secondary structures can form modular RNA motifs that can allow for small molecule recognition. (B) Structural schematic of microRNA processing. Primary transcripts (pri-miRNAs) are processed by the Drosha endonuclease to yield precursor hairpins (pre-miRNAs), which are exported to the cytoplasm and subsequently processed by the Dicer endonuclease to liberate a mature miRNA. One of the mature strands is then loaded into the argonaute/RNA-induced silencing complex (AGO/RISC), whereupon it acts on RNAs to modulate gene expression. Aberrant miRNA expression can be causative of disease phenotypes. (C) One rational design approach to target RNA is to understand the molecular recognition of structural elements by small molecules, those elements that are preferred by the small molecule and those that are discriminated against. Inforna compares structural elements within an RNA target to a database of these preferred interactions to afford lead small molecules. For example, binding to the pri- or pre-miRNAs at functional Drosha or Dicer sites can prevent their processing to the active, mature strand, thus allowing the rescue of disease-associated phenotypes through the inhibition of biogenesis.
