Figure 5.
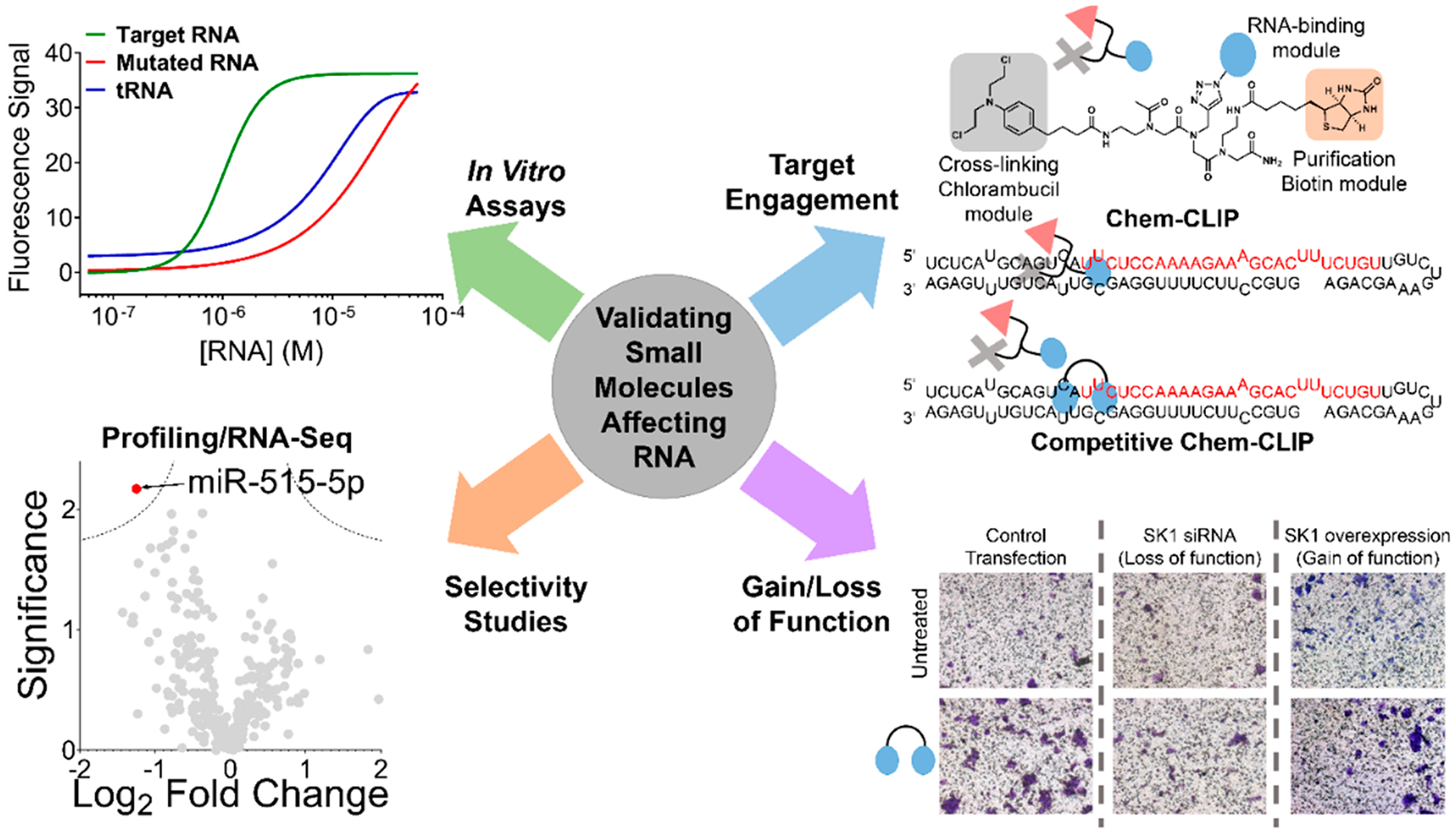
Tools to assess target engagement and selectivity of small molecules targeting RNA. Developing small molecules against RNA starts with identification of a hit, whether through Inforna (Figure 3) or screening approaches (target-based, phenotypic, fragment-based, DNA-encoded, etc.). Considering the factors from Figure 4, the hit must then be validated and optimized, including for in vitro binding affinity to the RNA structural element over RNAs that do not contain the motif and other abundant RNA/DNAs. Further validation in vitro and in cells can be accomplished with target engagement approaches that use chemical probing methods that measure RNA enrichment (Chem-CLIP) or RNA depletion (Competitive (C)-Chem-CLIP), among others. Comprehensively evaluating cellular selectivity on a transcriptome- and proteome-wide scale is also part of the workflow to validate a small molecule RNA target. After demonstration of selective on-target effects, the compound’s functional effect must then be validated in more advanced models, including the effect of a gain or loss in expression of the target.
