Figure 6.
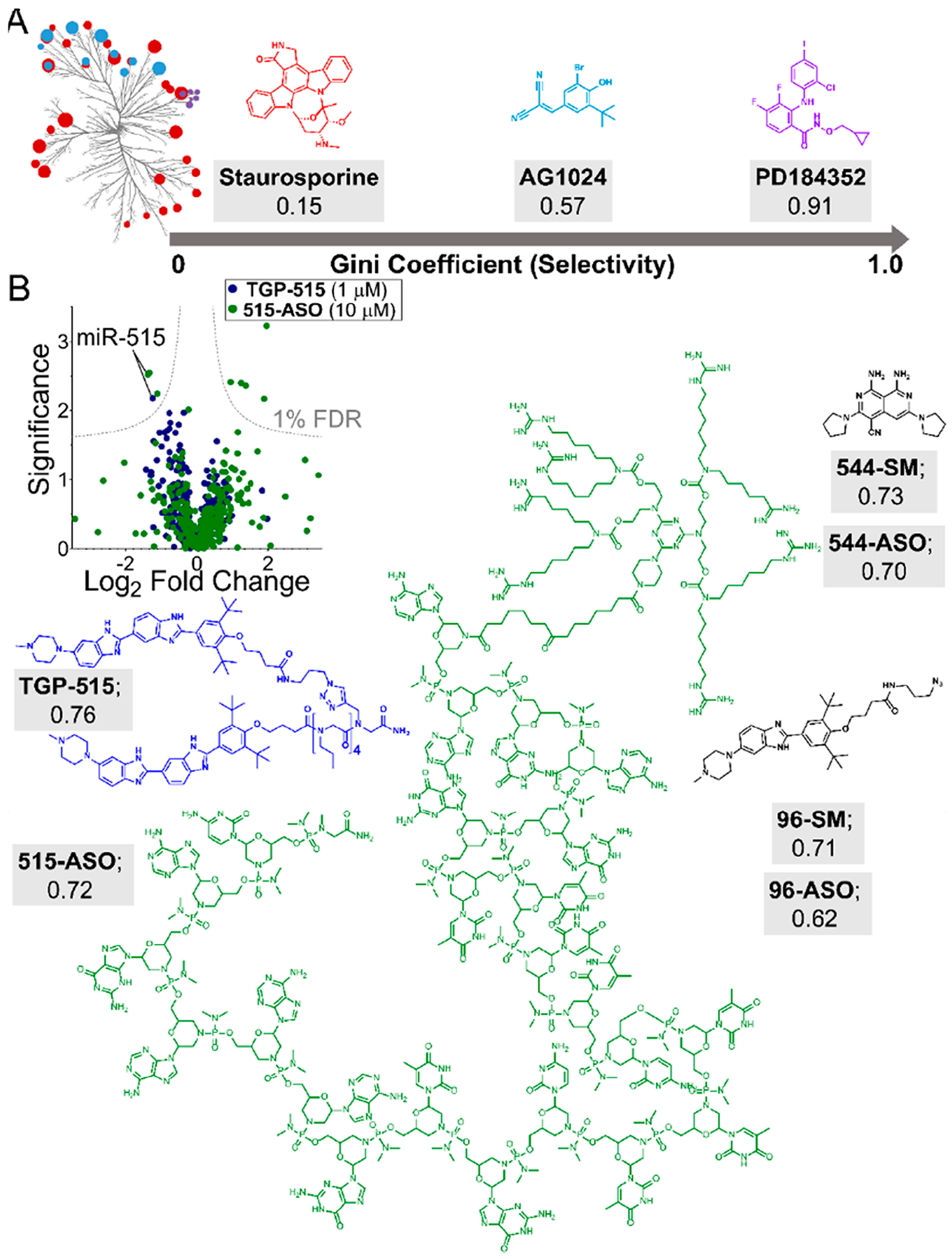
Quantitatively evaluating the selectivity of small molecules targeting RNA. (A) Data from profiling experiments can be used to quantify compound selectivity by calculating a Gini coefficient (GC). GC analysis of kinase inhibitors showed that promiscuous compounds (staurosporine) are characterized by values close to 0, while highly selective compounds exhibit Gini coefficient values close to 1 (PD184352; 0.91), with selective compounds being defined as >0.6. (B) GC analyses can be applied to profiling data (left), such as miRNA qPCR profiling data between small molecules (TGP-515; blue) and ASOs (515-ASO; green). Applying this analysis to various small molecule ligands targeting RNA structure indicates that they demonstrate high selectivity for their targets. Antisense oligonucleotides targeting RNA sequence are also selective for their targets. Shown are the structures and GC values of TGP-515 (blue) and Vivo-Morpholino ASO targeting miR-515 (green), in addition to other small molecule/ASO GC analyses. Overall, Gini coefficients provide a metric to quantitatively define compound selectivity. When applied to RNA targeting, GC analyses demonstrate that small molecules that recognize RNA structure can rival or exceed the selectivity of ASOs designed to bind via Watson–Crick base pairing.
