Figure 9.
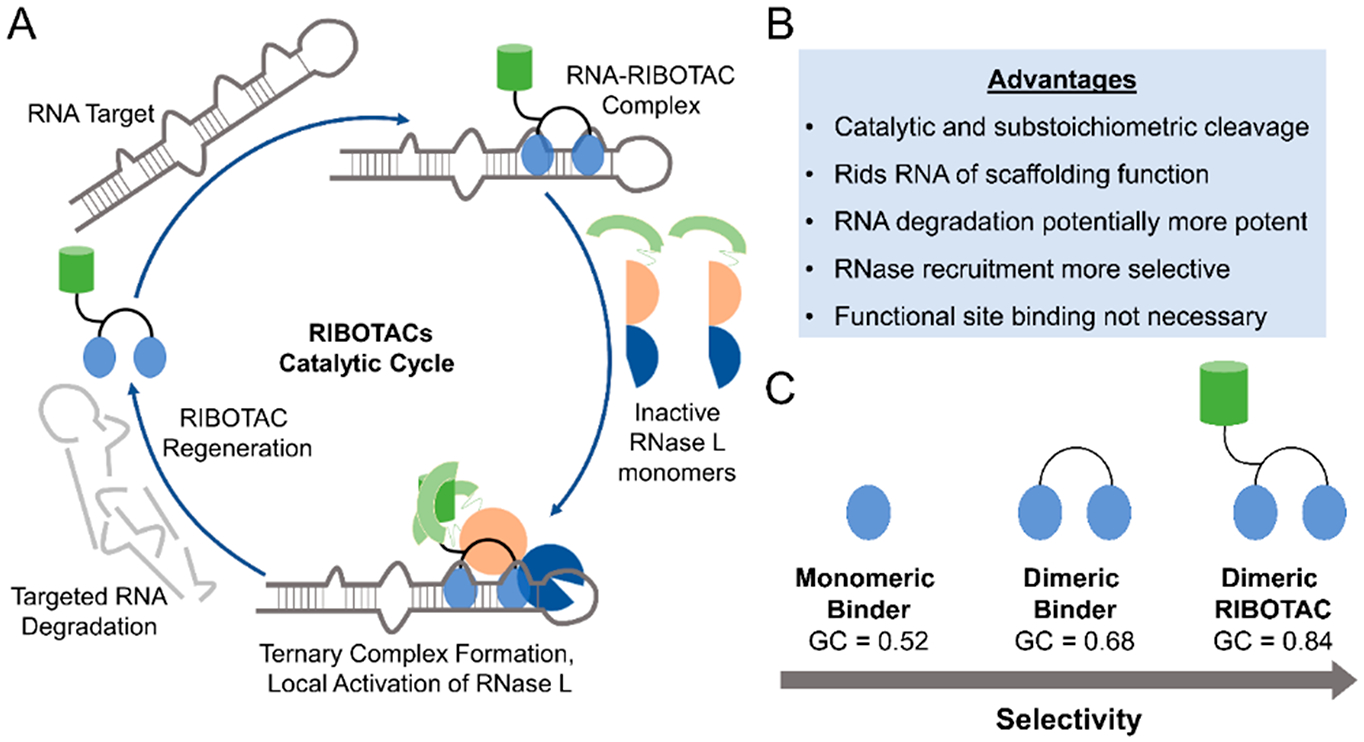
Ribonuclease targeting chimeras (RIBOTACs) as heterobifunctional degraders of RNA. (A) Taking cues from PROTACs and RNase H-based antisense oligonucleotide approaches, RIBOTACs are heterobifunctional compounds that recruit endogenous nucleases to degrade a targeted transcript. RIBOTACs can potentially increase potency of small molecules as they can catalytically and substoichiometrically degrade an RNA target. These RIBOTACs simply need to bind the target (not necessarily at a functional site) and use endogenous ribonuclease pathways to remove the RNA via targeted degradation, which also rids the RNA of any potential scaffolding functions with RBPs. Formation of the ternary complex may also increase selectivity as only meaningful interactions between the RNA:RIBOTAC:RNase L will result in cleavage. While this approach is potentially broadly applicable, development and optimization of both RNA binders and RNase-recruiting modules remain time-consuming, especially as there are a limited number of known RNase activators. Additionally, RIBOTACs that function through RNase L can have less pronounced effects on nuclear RNA, as RNase L is primarily cytoplasmic. (B) Advantages provided by the RIBOTAC approach. (C) Demonstration of increased selectivity of different RNA-binding modules, as indicated by GC analysis. The monomeric RNA-binding module that binds a single functional site on an RNA is less selective than the multivalent ligand targeting the same RNA. Adding an RNase L recruitment module to convert the dimeric compound into a RIBOTAC allows for increased selectivity, potentially due to the requirement of effective ternary complex formation between the RNA, RIBOTAC, and RNase L.
