Figure 3. Presentation of common TOP1 peptides linked to shared ATA-associated motifs in the HLA-DRβ chain.
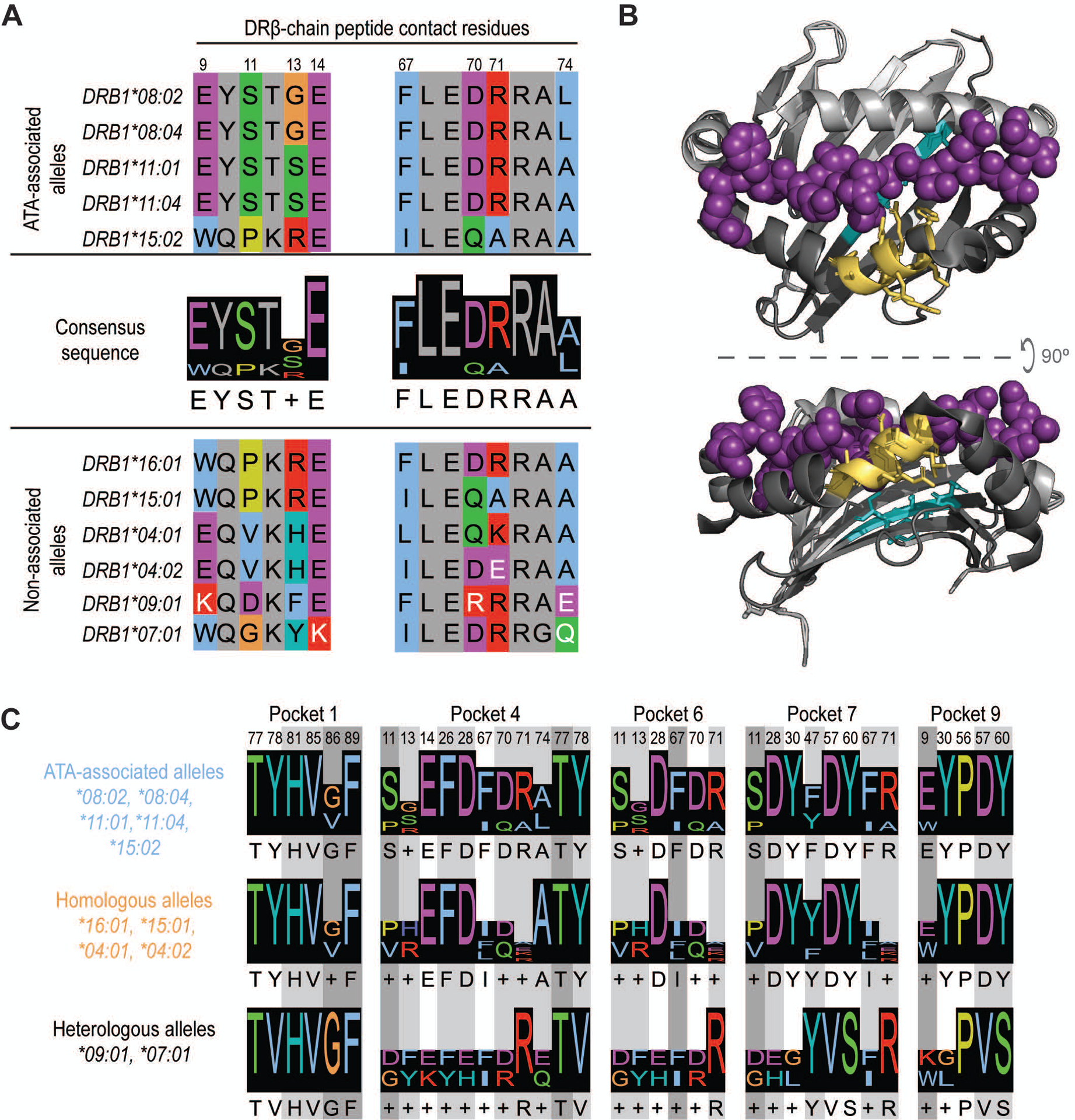
(A) Amino acid sequence alignment of ATA-associated (top panel) and non-associated alleles (bottom panel) is shown. The two consensus sequences identified in the peptide-binding groove of ATA-associated alleles are shown (middle panel). Peptide contact residues in the DRβ chain are shown (hydrophobic=blue, negative=purple, positive=red, polar=green, glycine=orange, prolines=yellow, and aromatic=cyan) and their position within the sequence is numbered, while intervening residues are shown in gray. The white text indicates non-conserved residues. (B) A depiction of HLA-DR11 crystalized with a peptide (6CPN) is shown and features of the peptide binding groove have been color-coded: DR α-chain residues (light gray), DR β-chain (dark gray) and peptide (magenta). The 9E-Y-S-T-S/G13 and 67F-L-E-D-R-R-A-A/L74 motifs are shown in cyan and yellow, respectively. (C) Peptide contact residues for each MHC binding pocket and the relevant consensus sequence for ATA-associated (light blue), homologous (orange) and heterologous (black) alleles are represented. Different shades of gray indicate the importance of each residue to peptide binding, with dark gray being the most critical.
