Figure 3 |. A surface-exposed 2-pyridyl moiety of CR8 confers glue degrader activity.
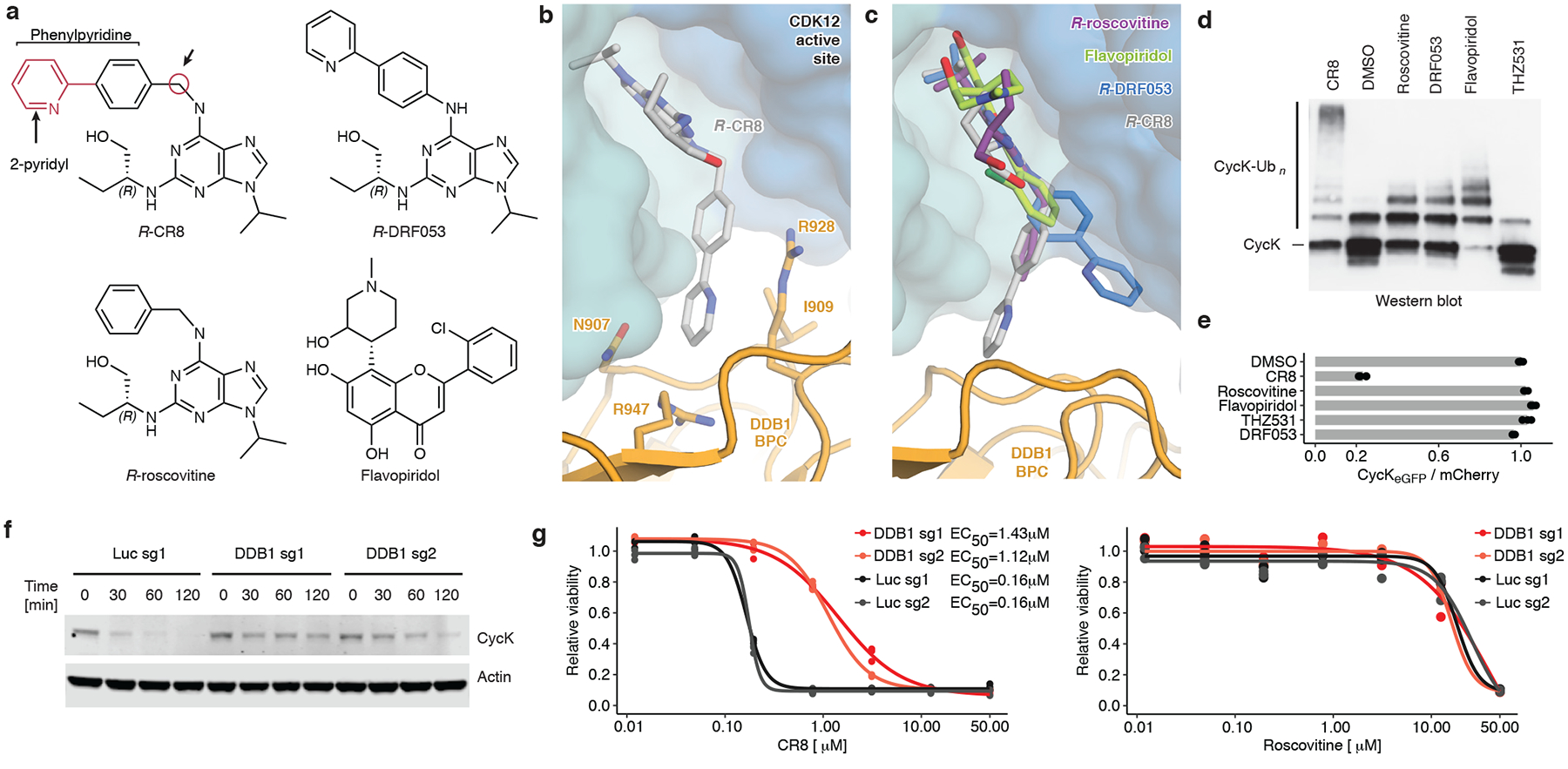
a, Chemical structures of CDK inhibitors. Arrows indicate differences between R-CR8, R-DRF053 and R-roscovitine. b, Close-up of the CDK12-CR8-DDB1 interface. The phenylpyridine moiety of CR8 contacts DDB1 residues. c, R-roscovitine (PDB entry 2A4L), R-DRF053 and flavopiridol (3BLR) in the active site of CDK12 in the DDB1-CR8-CDK12-cycK complex through superposition of kinase domains or the purine moiety (for DRF053). d, In vitro ubiquitination of CDK12-cycK complex by RBX1N8CUL4-DDB1 in the absence (DMSO) or presence of 2 μM compound (n=2). e, Flow analysis of CycKeGFP degradation in HEK293TCas9 cells treated with 1 μM compound for 2 hours (n=3). f, Immunoblots of CycK in HEK293TCas9 cells transfected with the indicated sgRNAs and treated with 1 μM CR8 (n=2). g, Drug sensitivity of sgRNA-transfected HEK293TCas9 cells after a 3-day exposure to CR8 or roscovitine (n=3).
