Figure 3. Cell type-specific analysis of gene expression changes in striatal spiny projection neurons (SPNs), astroglia, and cholinergic interneurons of the allelic series knockin models of HD by TRAP.
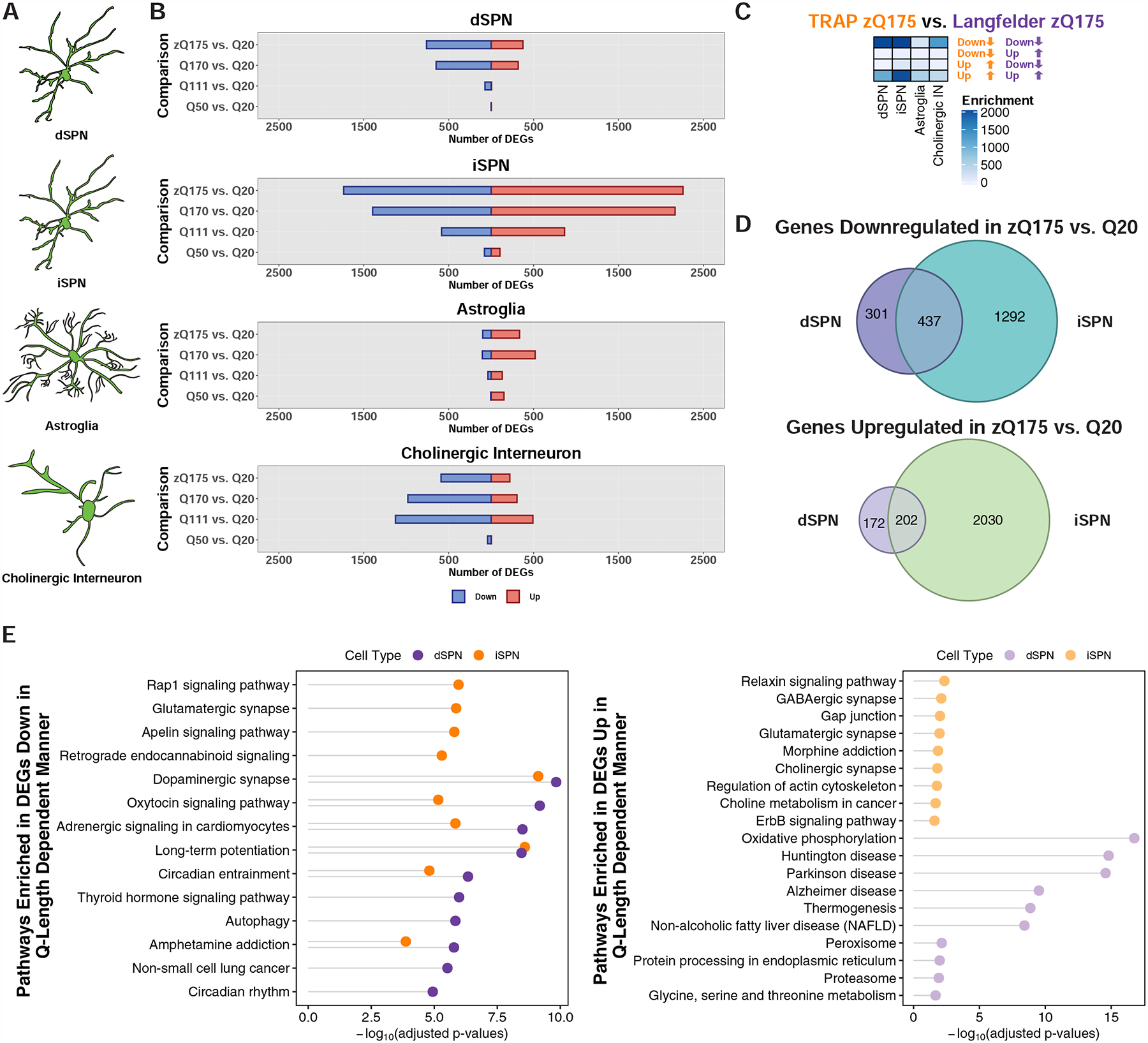
(A) Schematic depicting the targeting of striatal dSPNs, iSPNs, astroglia, and cholinergic interneurons. (B) Number of genes that were differentially expressed at the translated mRNA level in each of the cell types indicated at 6-months of age. (C) Overlap of the zQ175DN model downregulated and upregulated genes identified by cell type-specific TRAP and bulk RNA-Seq from the cited Langfelder et al. study. Overlaps were assessed using Fisher’s exact test and −log10-adjusted p-values are reported in the heatmap. (D) Venn overlap analysis of differentially expressed genes in the zQ175DN vs. Q20 comparisons between dSPNs and iSPNs (top panel: downregulated; bottom panel: upregulated). (E) KEGG pathway enrichment analysis of genes downregulated (left) and upregulated (right) in dSPNs and iSPNs from the linear regression analysis of CAG-length dependent changes, represented with Fisher’s exact test −log10-adjusted p-value.
