Figure 4. Cell type-specific analysis of gene expression changes in striatal cell types of the R6/2 and zQ175DN models of HD by single nuclear RNA-Sequencing (snRNA-Seq).
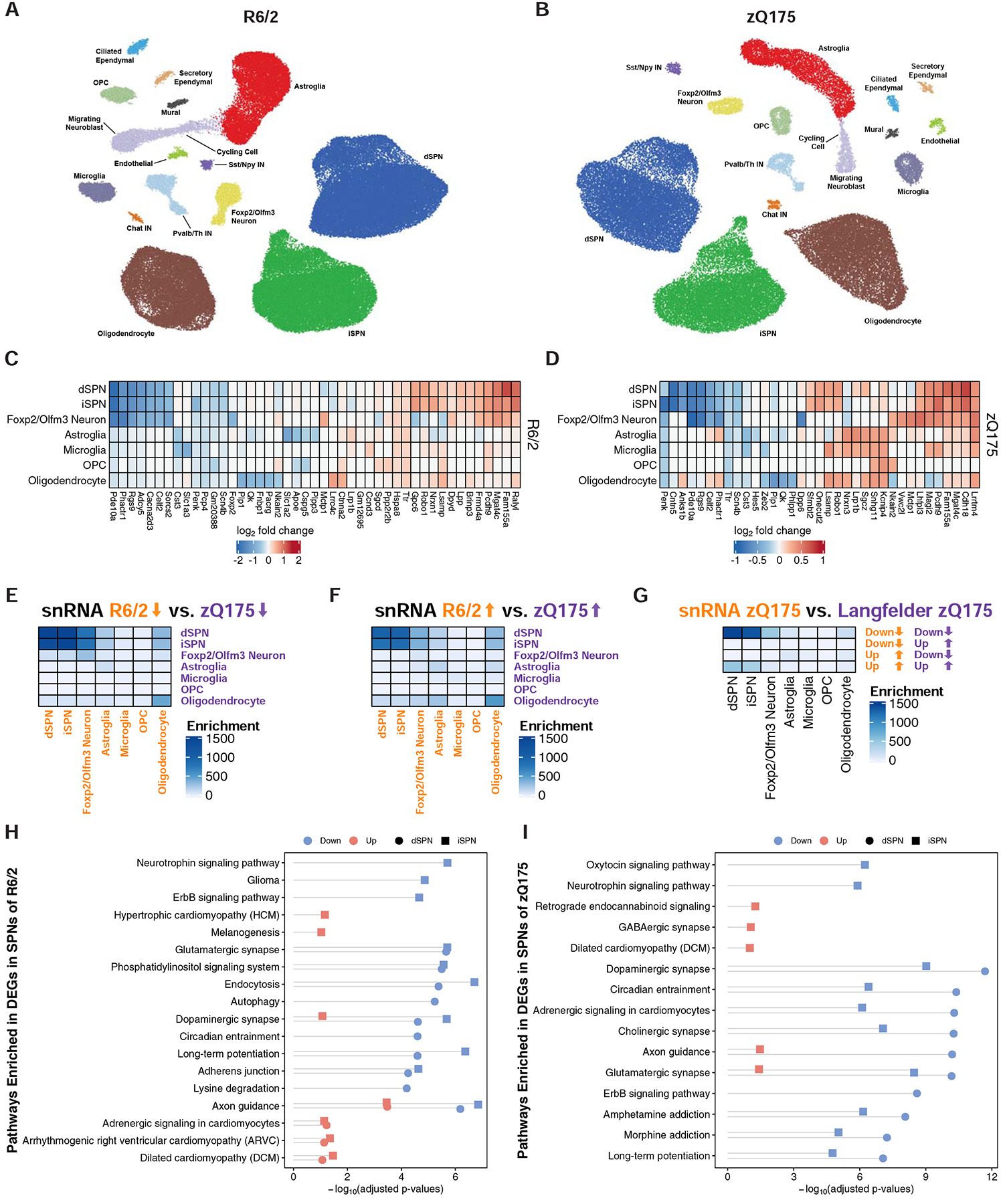
(A-B) Two-dimensional ACTIONet graphs of the major annotated cell types in the R6/2 (A) and zQ175DN (B) mouse models of HD (n = 108,467 nuclei across fifteen mice: eight isogenic control and seven R6/2 model mice, all at 9-weeks of age; n = 50,643 nuclei across eight mice: four isogenic control and four zQ175DN model mice, all at 6-months of age). Top five most downregulated and upregulated protein-coding genes by log2-fold change in the most abundant striatal cell types in the R6/2 model vs. control (C) or zQ175DN vs. control (D) comparisons. Significance of the overlap between cell type-specific (E) downregulated and (F) upregulated genes of snRNA-Seq-identified cell types across mouse models (Fisher’s exact test −log10-adjusted p-value). (G) Significance of overlap between downregulated and upregulated genes identified by snRNA-Seq and bulk RNA-Seq from the cited Langfelder et al. study in the zQ175DN model. The overlaps were assessed using Fisher’s exact test and −log10-adjusted p-values are reported. KEGG pathway analysis of downregulated and upregulated genes in dSPNs and iSPNs in the R6/2 (H) and zQ175DN (I) models, represented with Fisher’s exact test −log10-adjusted p-value.
