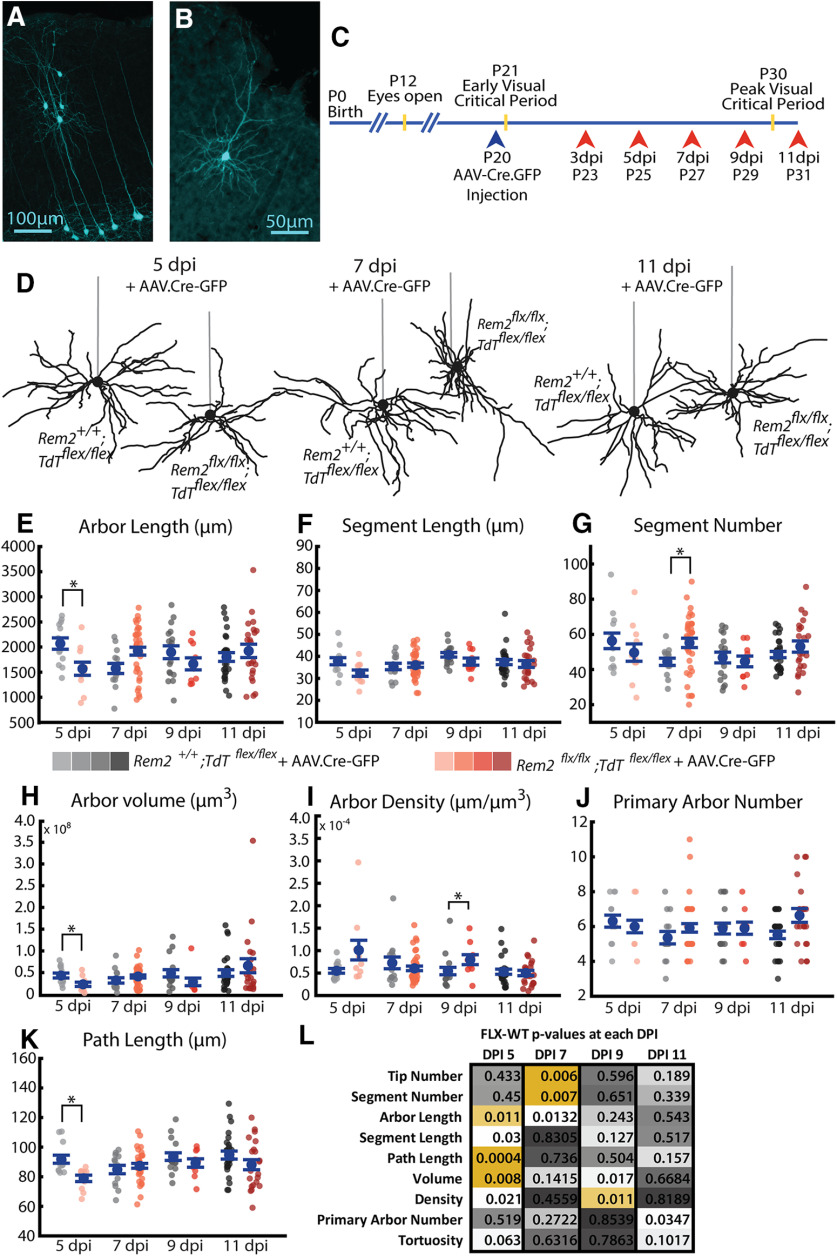
Figure 10.
Acute deletion of Rem2 results in many transient changes in basal arbor architecture. A, Example sparse population of WT neurons infected with AAV.Cre-GFP in primary visual cortex expressing Ai9.TdTomato reporter (pseudocolored cyan). B, Example single WT neuron expressing Ai9.TdTomato reporter (pseudocolored cyan). Entire dendritic arbor is visible and amenable to reconstruction. C, Timeline for AAV.Cre-GFP injection (blue arrowhead) and morphologic sample collection schedule (coral arrowheads). D, Example L2/3 pyramidal neurons in the primary visual cortex of Rem2+/+;Tdtflex/flex + AAV.Cre-GFP and Rem2flx/flx;Tdtflex/flex + AAV.Cre-GFP mice sampled 5, 7, or 11 dpi. Straight gray lines diagram the unreconstructed apical dendrite to indicate the orientation of cells. E, Length of the complete basal arbor of Rem2+/+;Tdtflex/flex + AAV.Cre-GFP and Rem2flx/flx;Tdtflex/flex + AAV.Cre-GFP neurons sampled at 5, 7, 9, and 11 dpi. F, Average segment length of neurons from Rem2+/+;Tdtflex/flex + AAV.Cre-GFP and Rem2flx/flx;Tdtflex/flex + AAV.Cre-GFP mice sampled at 5, 7, 9, and 11 dpi. G, Segment number of neurons from Rem2+/+;Tdtflex/flex + AAV.Cre-GFP and Rem2flx/flx;Tdtflex/flex + AAV.Cre-GFP mice sampled at 5, 7, 9, and 11 dpi. H, Volume of the complete basal arbor of Rem2+/+;Tdtflex/flex and Rem2flx/flx;Tdtflex/flex + AAV.Cre-GFP neurons sampled at 5, 7, 9, and 11 dpi. I, Density of the complete basal arbor of Rem2+/+;Tdtflex/flex + AAV.Cre-GFP and Rem2flx/flx;Tdtflex/flex + AAV.Cre-GFP neurons sampled at 5, 7, 9, and 11 dpi. J, Number of basal dendritic primary arbors of Rem2+/+;Tdtflex/flex + AAV.Cre-GFP and Rem2flx/flx;Tdtflex/flex + AAV.Cre-GFP neurons sampled at 5, 7, 9, and 11 dpi. K, Average path length of neurons from Rem2+/+;Tdtflex/flex + AAV.Cre-GFP and Rem2flx/flx;Tdtflex/flex + AAV.Cre-GFP mice sampled at 5, 7, 9, and 11 dpi. L, Summary of p values from all comparisons between Rem2+/+;Tdtflex/flex + AAV.Cre-GFP and Rem2flx/flx;Tdtflex/flex + AAV.Cre-GFP neurons at each of the sampled dpi time points with significant results shown in yellow. Arbor characteristics impacted by Rem2 deletion are not conserved across ages/time points. Error bars indicate mean ± SEM. Blue dots indicate means. Gray/coral dots indicate single neurons. For all Rem2flx/flx;Tdtflex/flex + AAV.Cre-GFP measurements: 5 dpi: N = 11 cells, 4 mice; 7 dpi: N = 38 cells, 7 mice; 9 dpi: N = 10 cells, 4 mice; 11 dpi: N = 22 cells, 4 mice. For all Rem2+/+;Tdtflex/flex + AAV.Cre-GFP measurements: 5 dpi: N = 13 cells, 5 mice; 7 dpi: N = 14 cells, 5 mice; 9 dpi: N = 17 cells, 4 mice; 11 dpi: N = 27 cells, 6 mice. *p < 0.0125 (Wilcoxon rank-sum test, Bonferroni-corrected α = 0.0125).