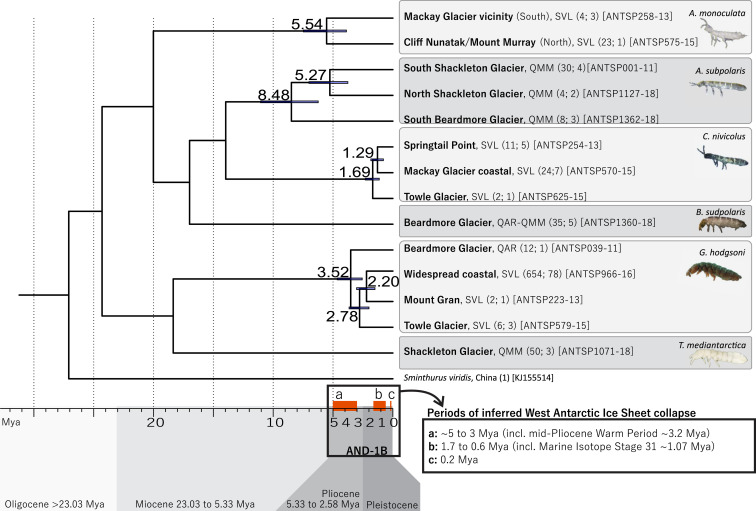
Fig. 5.
Dating estimates for the four springtail species with high intraspecific variability (>3.3% divergence) based on the molecular clock rate of 0.0168 (i.e., 3.54% divergence per million years; ref. 37). Estimated dates (in millions of years) of population isolation, and bars representing 95% highest posterior density, are shown at nodes (see also Table 2). Based on the AND-1B core data (3) and simulated modeling (4), collapse of the WAIS likely occurred multiple times during three major periods within the last 5 My (shown as orange boxes on the timeline), facilitating connectivity of Collembola populations. Number of sequences and unique haplotypes, respectively, are shown in parentheses, followed by the BOLD sequence ID of the exact sequences (658 bp) used for molecular clock estimations.