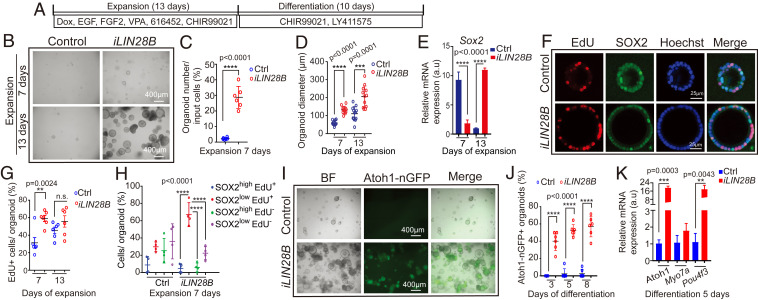
Fig. 2.
LIN28B overexpression promotes cochlear epithelial cell expansion and hair cell production. Cochlear organoid cultures were established from stage P5 Atoh1-nGFP;iLIN28B transgenic mice and control littermates that lacked the LIN28B transgene. Atoh1-nGFP marks nascent hair cells. (A) Experimental strategy. (B) BF images of control and LIN28B overexpressing (iLIN28B) organoid cultures at 7 and 13 d of expansion. (C) Organoid forming efficiency in control (Ctrl, blue) and LIN28B (iLIN28B, red) overexpressing cultures (n = 6, two independent experiments). (D) Organoid diameters in control (Ctrl, blue) and LIN28B (iLIN28B, red) overexpressing cultures after 7 and 13 d of expansion (n = 12, two independent experiments). (E) qRT-PCR analyzing Sox2 mRNA expression in control (Ctrl, blue bar) and LIN28B overexpressing (iLIN28B, red bar) organoids at 7 and 13 d of expansion (n = 3, from one representative experiment, three independent experiments). (F) Cell proliferation in control and LIN28B overexpressing organoids. An EdU pulse was given at 7 d (shown) or 13 d of expansion and EdU incorporation (red) was analyzed 2 h later. SOX2 (green) marks supporting cells/prosensory cells, Hoechst labels cell nuclei (blue). (G) Percentage of EdU+ cells in control (Ctrl, blue) and LIN28B overexpressing (iLIN28B, red) organoids at 7 and 13 d of expansion (n = 6, two independent experiments, n.s. not significant). (H) EdU incorporation in SOX2-high and SOX2-low expressing cells (n = 4, two independent experiments). Note that the majority of EdU+ cells in LIN28B overexpressing organoids expressed SOX2 at a low level (red, SOX2low EdU+). (I) BF and green fluorescent (Atoh1-nGFP) images of control and LIN28B overexpressing organoids after 8 d of differentiation. (J) Percentage of Atoh1-nGFP+ organoids in control (Ctrl) and LIN28B overexpressing (iLIN28B) cultures after 3, 5, and 8 d of differentiation (n = 6, two independent experiments). (K) qRT-PCR analyzing hair cell-specific mRNA expression (Atoh1, Myo7a, Pou4f3) in control (Ctrl, blue bar) and LIN28B overexpressing (iLIN28B, red bar) organoids after 5 d of differentiation (n = 3, from one representative experiment, three independent experiments). Bars in E and K represent mean ± SD, otherwise individual data points and their mean ± SD were plotted. Individual data points in D, G, and H represent the average value per animal. n = animals analyzed per group. Two-tailed, unpaired Student’s t test was used to calculate P values. a.u., arbitrary unit.