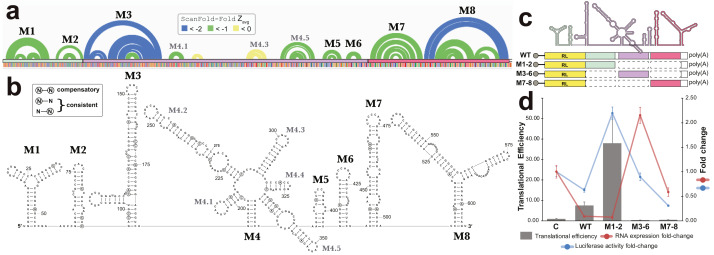
Figure 6. Results for the region partially overlapping BFRF1-3.
(A) The top of this panel shows ScanFold-Fold predicted base pairs represented as colored arcs. (B) Secondary structure models predicted after energy minimization using ScanFold-Fold base pairs (average z-score < − 1) as constraints. Eight motifs (M1-8) were defined (labeled in both the (A) and (B). Nucleotides with mutations preserving the shown base pair are circled on the structure models. (C) Cartoon showing the locations of three regions (colored green, purple, and red) that were added to downstream of Renilla luciferase (RL; diagrams of constructs are shown with RL in yellow and the added fragments of EBV-1 in colors corresponding to the cartoon). (D) Results of luciferase assays for each construct; the translational efficiency and luciferase mRNA and protein fold-change (vs. empty vector control (C)) are plotted for C, wild type (WT) and three fragments of the structured region. The translational efficiencies reported here were calculated by dividing the relative protein abundance of RL (RRR values) by mRNA levels of RL (2−ΔΔCT values); see methods for detail and Table S2 for raw values.