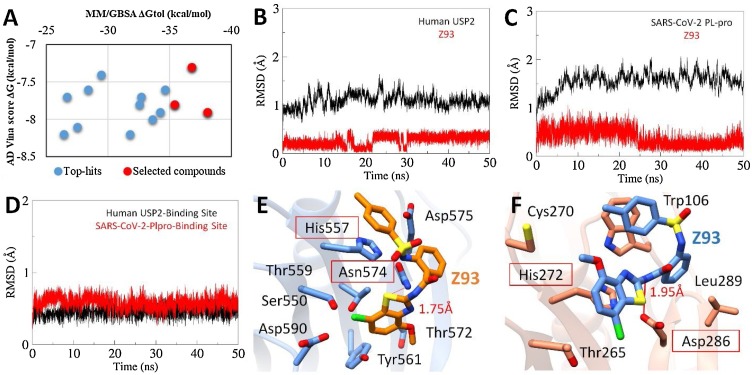
Fig. 5.
(A) The energy plot of top hits between corresponding AD Vina docking score and MMGBSA total binding free energy calculated after a production run of 20 ns MD simulations). The selected compounds are highlighted red. Root-mean-square-deviation of USP2 (B) and SARS-CoV-2 PLpro (C) backbone are estimated over a period of 20 ns with bound Z93 ligand (red). (D) RMSD plot of binding site residues of corresponding proteases. The MD simulated binding modes of Z93 (sticks representation) inside the active site of USP2 (E) and SARS-CoV-2 PLpro (F).