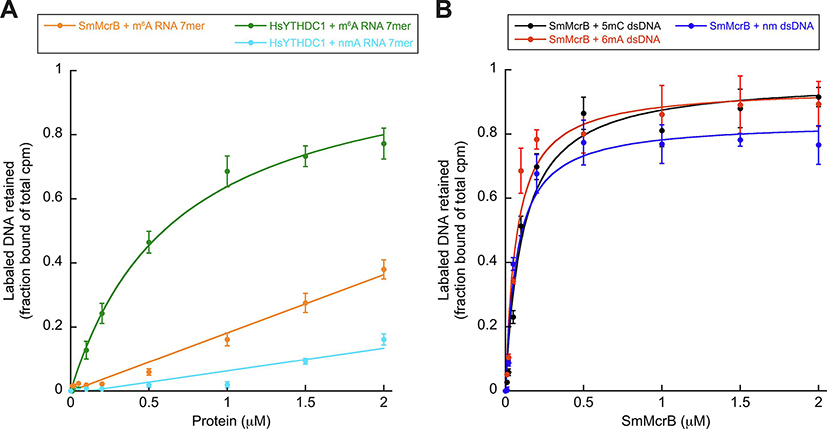
Figure 4. SmMcrB preferentially binds DNA and is promiscuous.
All data points represent average of three independent experiments (mean ± standard deviation) using multiple, independently purified batches of protein. Binding constants were determined by nonlinear curve fitting using Kaleidagraph (Synergy Software) and defined as the concentration of the protein at which 50% of the labeled DNA substrate is retained. Substrate sequences and calculated Kd values are listed in Supplementary Table 2 and Table 1 respectively. Abbreviations are as follows: ds, double stranded; nm, non-methylated; m6A, 6-methyladenine- modified; m5C, 5-methylcytosine-modified. A. Filter binding analysis of Sm3–180 and HsYTHDC1 YTH domain interactions with RNA containing the YTH consensus sequence. B. Filter binding analysis of Sm3–180 interactions with dsDNA substrates.