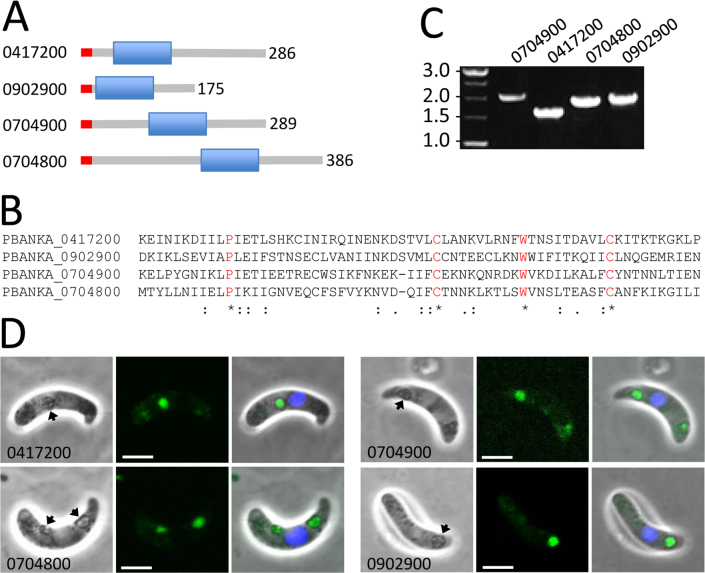
Fig. 2.
Characterization of pleckstrin homology-like (PHL) domain proteins PBANKA_0417200, PBANKA_0704800, PBANKA_0704900 and PBANKA_0902900. A: Schematic diagram of protein structures with relative positions of the PHL domains and ER signal peptides (red). Protein lengths (amino acids) are indicated on the right-hand side. B: Alignment of the shared domain reveals a unique amino acid signature C(X)9W(X)9C (one letter amino acid code, X = any amino acid). C: PCR diagnostic for integration of the GFP-tagged alleles into the target loci gives rise to the expected products of approximately 1.6 kb (PBANKA_0417200, primers PH1–5’diag (TTATATAATAAATCCTAACACTTCATCG) and GFP-R (GTGCCCATTAACATCACC)); 1.9 kb (PBANKA_0704800, primers PH2–5’diag (AAAGTATGAACGCATTAAAAAAATC) and GFP-R); 2.1 kb (PBANKA_0704900, primers PH3–5’diag (TACAGGTAAAAAAGATTGGCAT) and GFP-R); and 2.0 kb (PBANKA_0902900, primers PH3–5’diag (CGATTTTACATTTACTATTTTGTTAAAAAG) and GFP-R). D: Brightfield and fluorescence confocal images of ookinetes expressing GFP-tagged PBANKA_0417200, PBANKA_0704800, PBANKA_0704900 and PBANKA_0902900, showing localisation in focal spots associated with pigment (arrows) characteristic of crystalloids. Hoechst DNA staining (blue) labels nuclei. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)