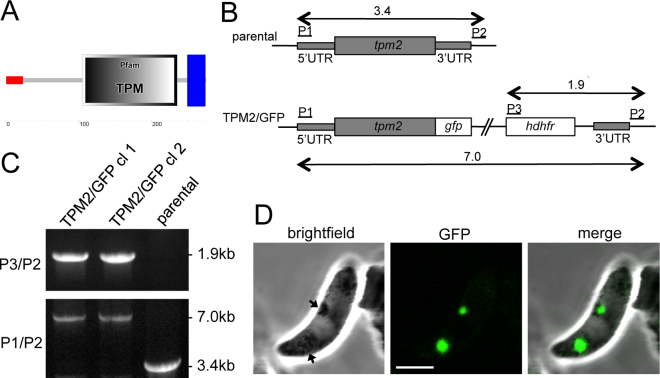
Fig. 4.
Characterization of TPM domain protein PBANKA_1104100 (TPM2). A: Predicted structure showing ER signal peptide (red), TPM domain and C-terminal transmembrane domain (blue), produced with Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de). B: Schematic diagram of the unmodified (parental) and modified tpm2 allele in parasite lines TPM2/GFP. The tpm2 gene is indicated with coding sequence (wide grey bars, introns not shown) and 5′ and 3′ untranslated regions (UTRs) (narrow grey bars). Also indicated are the relative positions of the GFP module (gfp); the human DHFR selectable marker gene cassette (hdhfr); and primers used for diagnostic PCR amplification (P1-P3). Primer P2 sequence is not present within the targeting vector. Sizes of PCR products are also indicated. C: Diagnostic PCR for integration into the target locus with primers P3 (ACAAAGAATTCATGGTTGGTTCGCTAAACT) and P2 (CATCTTGAGGTATTTGTGCATATTC), giving rise to a 1.9 kb product (top panel). Diagnostic PCR with primer pair P1 (ACGAAGTTATCAGTCGAGGTACCGCTCAAACTATTCCTCCTCAATC) and P3 amplified an approximately 3.4 kb fragment from the parental WT parasites, and a 7.0 kb fragment in the TPM2/GFP parasites, confirming absence of the unmodified allele in the transgenic parasite line (bottom panel). D: Brightfield and GFP fluorescence images of a live ookinete of parasite line TPM2/GFP, showing localisation in focal spots associated with pigment (arrows) characteristic of crystalloids. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)