Fig. 8.
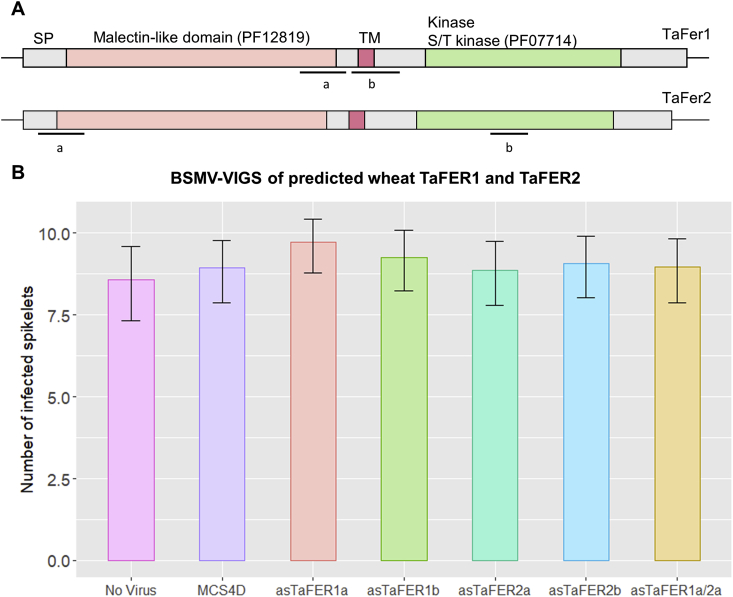
BSMV-VIGS of Feronia genes in wheat. A) Diagrammatical representation of wheat TaFer1 and TaFer2 protein sequence showing both predicted kinase-like (PF07714) and malectin-like (PF12819) protein domains. Bars extending from the termini of the predicted protein structures indicate nontranslated cDNA regions up- and downstream of the coding regions. Bars below each gene model indicate the a and b regions amplified to generate different Barley stripe mosaic virus-mediated virus-induced gene silencing (BSMV-VIGS) constructs targeting these genes. SP = signal peptide, TM = transmembrane region. B) Graph representing number of visibly diseased spikelets below the F. graminearum inoculated points in wheat spikes at 15 dpi. “No Virus” represents wheat plants with no virus inoculation prior F. graminearum infection. “MCS4D″ represents control virus treatment where the only addition to the viral genome is a multiple cloning site (MCS). The BSMV-VIGS silencing constructs include asTaFER1a, asTaFER1b, asTaFER1a/2a, asTaFER2a and asTaFER2b. This graph represents a total of three combined experiment. Treatments did not present statistically significant differences in number of diseased spikelets, relative to BSMV:MCS4D control (p > 0.05 from GLMM analysis).