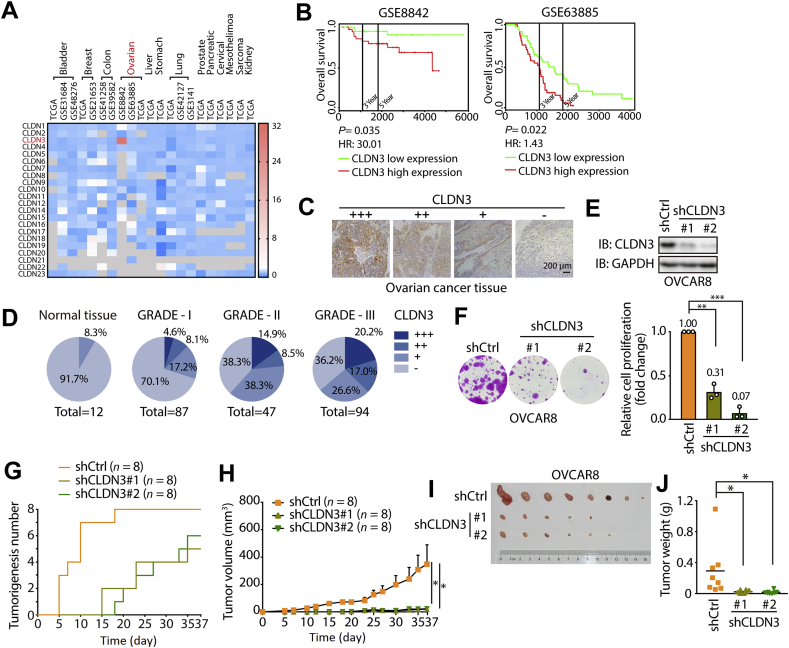
Figure 1.
Clinicopathologic characteristics of CLDN3 in ovarian cancer. (A) The HR (hazard ratio) value of each human CLDN family protein (data of 22 CLDNs are available) expression in different types of cancers. All data is obtained from TCGA and GEO dataset. The threshold of scale bar is from 0 to 32 and white color indicates the baseline value of 2. Shades of gray indicate no available data. (B) Kaplan–Meier curves for overall survival rate of patients with ovarian cancer from the GEO dataset (GSE8842 and GSE63885; green curve, patients with low expression of CLDN3; red curve, patients with high expression of CLDN3). (C) Representative images of different score of immunohistochemical stain, indicating different expression level of CLDN3 (+++ indicates high expression; ++ indicates median expression; + indicates low expression; – indicates negative expression). (D) The pie chart shows the proportion of different expression level of CLDN3 in each ovarian tumor grade (from GRADE-1 to GARDE-III and normal tissue group). Darker shades of blue indicate higher expression of CLDN3 (n = 240). (E) The knockdown efficiency of shRNA against CLDN3 in OVCAR8 cells was validated by immunoblotting. (F) Colony forming assay was performed on shCtrl, shCLDN3#1 and shCLDN3#2 OVCAR8 cells. Quantification of colony numbers relative to shCtrl group are shown on the right panel. Values with error bars indicate mean ± SD of three biological replicates (∗∗P < 0.01; ∗∗∗P < 0.001). (G) The tumorigenesis of shCtrl, shCLDN3#1 and shCLDN3#2 groups according to time course (37 days). The first time that tumor was found in each group was on day 5, day 15 and day 18. In shCtrl group, 8 tumors formed on day 18 which remained to the end of in vivo assay. In shCLDN3#1 group, 4 tumors formed on day 23 which remained to the end of in vivo assay. In shCLDN3#2 group, 6 tumors formed on day 35 which remained to the end of in vivo assay. (H) Tumor growth curves of shCtrl, shCLDN3#1 and shCLDN3#2 groups in vivo animal assay. Error bars represent SEM. (∗P < 0.05). (I) Tumor formed in the nude mice at the end of in vivo assay (n = 8 for each group). At the endpoint of the in vivo animal assay, the tumorigenesis ratio (the number of tumorigenic individuals/the number of total individuals) of each group: 8/8 in shCtrl group, 5/8 in shCLDN3#1 group, 6/8 in shCLDN3#2. The tumors were arranged from left to right by weight. (J) The nude mice were sacrificed, and the tumors were removed and measured in weight. Dot plots represent the weight of tumors (n = 8 for each group in all statistical analysis; ∗P < 0.05).