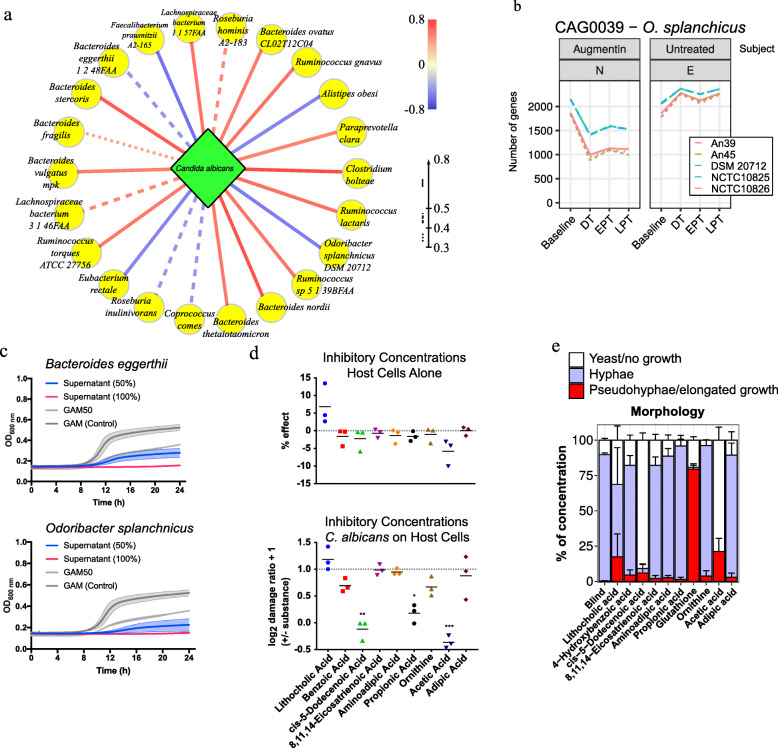
Fig. 4.
Candida albicans growth promotors and inhibitors. a Bacterial species co-abundant with C. albicans. Line colours and type indicate correlation coefficients. b Example using Odoribacter splanchnicus for genomic strain inference from metagenomic species (MGS) reads. Strains were inferred for each time point (x-axis) from number of genes with 0.5 reads per base (y-axis) per-reference genome. Data are from the antibiotic-treated participant N and untreated subject E. The number of genes coverage for each tested O. splanchnicus strains are shown. DSM 20712 was identical with another strain labelled NCTC10825. c Growth rate inhibition of C. albicans strain SC5314 cultivated with 50% and 100% bacterial supernatant (from Bacteroides eggerthii and Odoribacter splanchnicus) compared to control of medium (mGAM) only. d Damage of human vaginal epithelial cells (A431) based on release of lactate dehydrogenase (LDH) with metabolites at inhibitory concentrations. Grey lines, zero effect. Positive values imply cell damage. (Top) Human cells cultured without C. albicans. Effect compared to untreated cells. (Bottom) Cells co-cultured with C. albicans. Values are relative to damage caused by C. albicans without additional metabolites. Negative values imply less cell damage. e Composition of morphology of C. albicans cultures quantified by concentration of morphological types. Some metabolites cause atypical formation of hyphae-like structures (“Pseudohyphae/elongated yeast chains”). Some inhibited the formation of filaments or disrupted growth in general (“Yeast/no growth”)