Figure 2. Cancer cell migration in MDSC –PyMT cancer cell polydispersed co-culture on patterned PDMS substrates.
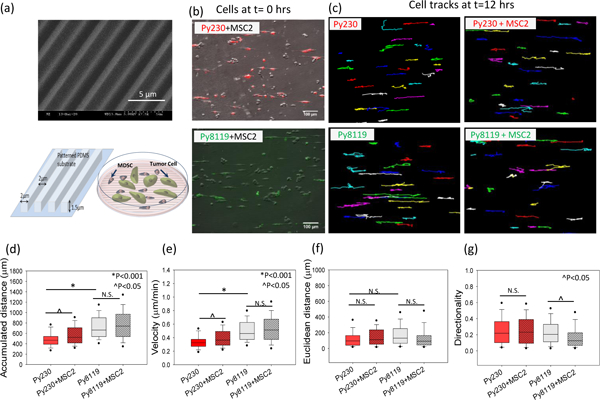
(a) Scanning electron micrograph (SEM) and schematic diagram of patterned PDMS substrates and cell migration setup for PyMT mouse cancer epithelial (Py230) and mesenchymal (Py8119) cells in polydispersed co-culture with mouse MDSCs (MSC2) on these substrates. For the migration studies, (b) tumor cells were labeled with live cell tracker fluorescent dye to differentiate them from the MSC2 cells in co-culture and (c) individual tumor cells were tracked over 24 hours to evaluate migration parameters. (d, e) A significant increase in accumulated distance and cell velocity was observed for the Py230 cells in the presence of MSC2s, while no significant difference was found in cell velocity for mesenchymal cells (Py8119) under co-culture conditions compared to monoculture. (f, g) The Euclidean distance and migration directionality do not change for Py230 cells while the directionality drops significantly for the Py8119 cells in the presence of MSC2 cells. One Way ANOVA on ranks was used to establish p-value. (n=3 samples, 60–100 cells per group), scale bar =100μm.
