Figure 2.
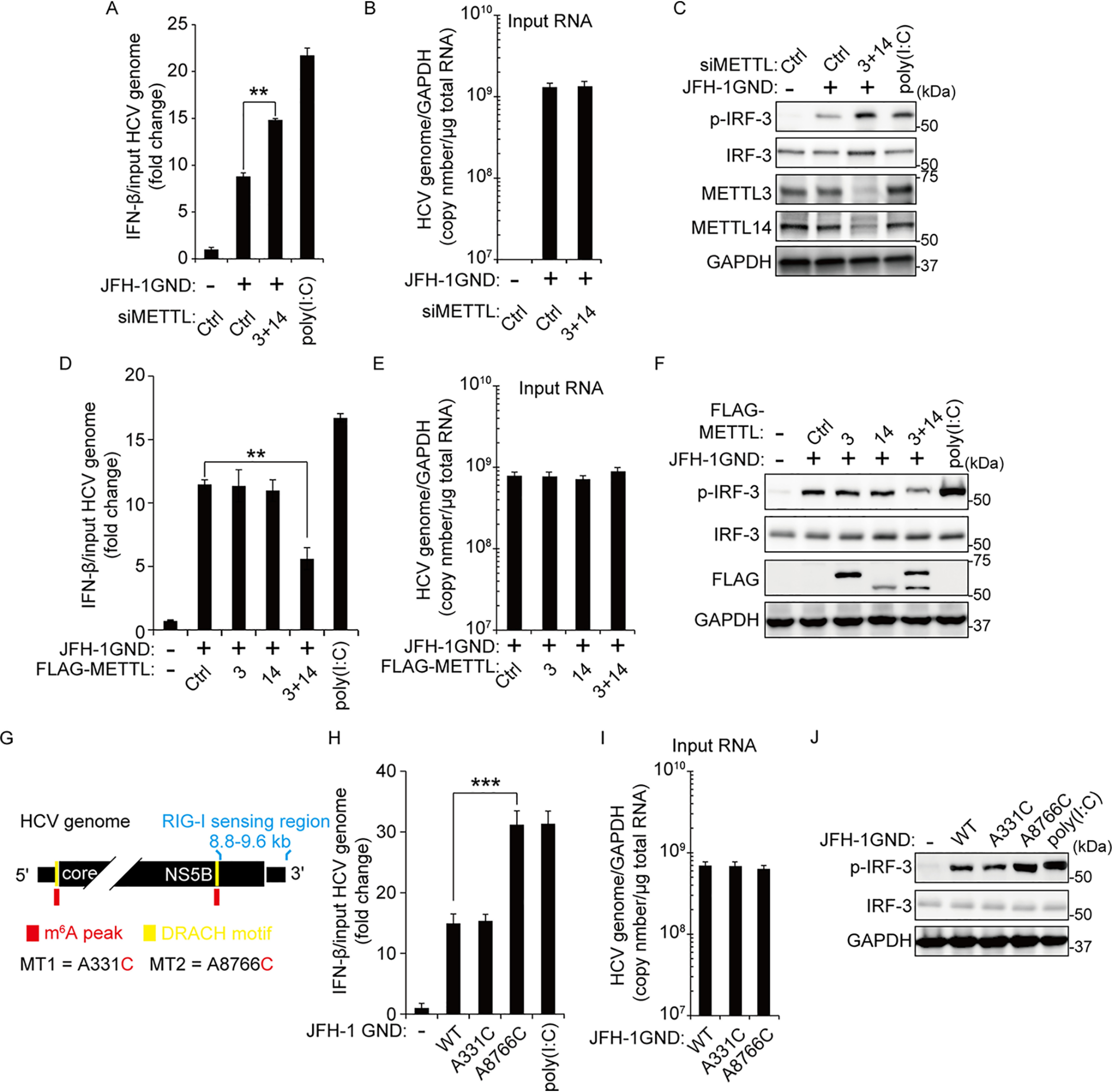
The m6A modification of HCV genome inhibits IRF-3–mediated IFN signals. A–C, Huh7 cells treated with siMETTL3 + 14 or control siRNA were transfected with JFH-1 GND RNA. IFN-β mRNA levels were assessed by qRT-PCR at 16 h post–JFH-1 GND RNA transfection. The HCV genome titers were analyzed by qRT-PCR for normalizing IFN-β mRNA (B). The p-IRF-3 protein levels were analyzed by immunoblotting in whole-cell lysates (C). D–F, FLAG-METLL3/14 transfected with Huh7 cells were treated with JFH-1 GND RNA for 16 h. The cells were harvested to assess expression levels of IFN-β mRNA (D) or the indicated proteins (F). The input HCV genome levels were quantified for calculating IFN-β mRNA (E). G, schematics indicate the location of the A8766C mutation of the m6A site in the NS5B-3′-UTR region of the HCV genome. The A331C mutation is in domain IV of the HCV IRES region. Red indicates m6A peak by MeRIP analysis. Yellow indicates the DRACH motif in the m6A peak. H–J, qRT-PCR analysis of IFN-β mRNA relative to input HCV genome in JFH-1 GND RNA or JFH-1 GND A8766C RNA transfected with Huh7 cells. The cells were harvested at 16 h post-transfection, and the relative levels of IFN-β mRNA (H) or p-IRF-3 (J) were analyzed. The input JFH-1 GND RNA levels were analyzed by qRT-PCR (I). JFH-1 A331C RNA was used as a control. The data for this figure are from three independent experiments, and the bars represent the means ± S.D. Statistical significance of the difference between groups was determined via an unpaired Student's t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Ctrl, control.
