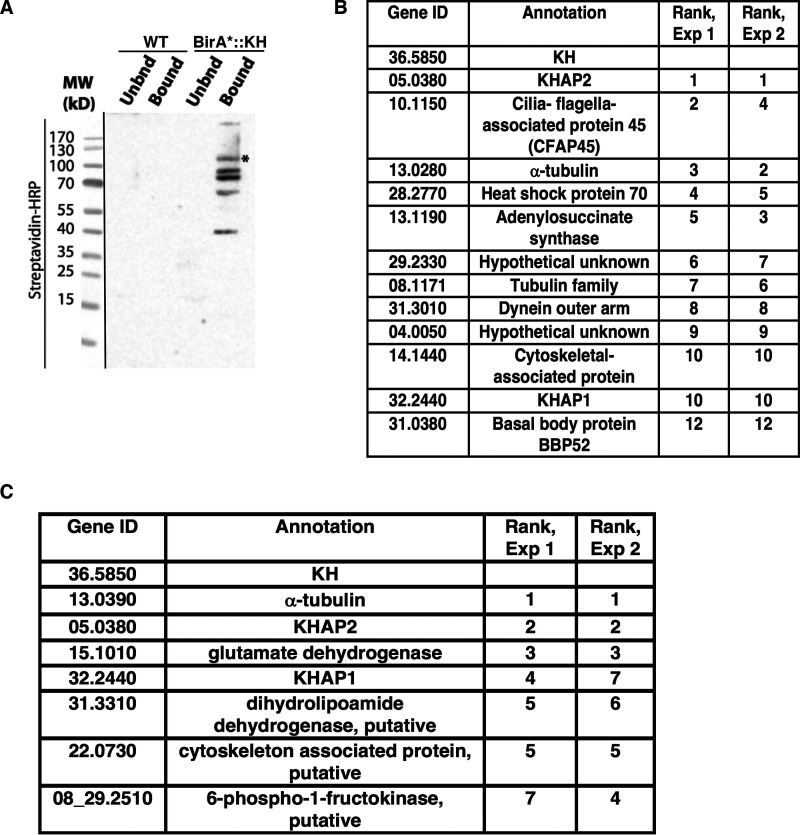
Figure 1.
Identification of candidate KH-interacting proteins. A, proximity biotinylation with BirA*-tagged KH. Both WT L. mexicana promastigotes (WT, not expressing BirA*) and promastigotes expressing BirA*::KH (BirA*-KH) were incubated with biotin, and lysates were subjected to affinity chromatography and eluted from streptavidin-agarose resin. Equivalent fractions of both resin-unbound (Unbnd) and bound and eluted (Bound) material were separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and probed with streptavidin-horseradish peroxidase (Streptavidin-HRP). The asterisk (*) band represents the BirA*::KH bait protein. The panel on the left (separated by a black line) shows a reference image of the protein ladder. B, BioID hits for BirA*::KH. Two experiments were performed, and spectral counts from WT samples were subtracted from counts from BirA*::KH-expressing promastigotes. Proteins that showed the largest differential in spectral counts in each of the two experiments are listed, with their relative rank in each experiment indicated. For simplicity, gene IDs are listed without the prefix LmxM used in the TriTrypDB genome database. Proteins with a difference larger than 10 spectral counts in both experiments are included in this list, with the addition of the protein corresponding to gene ID 31.0380, listed despite the differential count value of <10 for Experiment 2, due to its potential interest as a likely basal body protein. More detailed experimental results are reported in Table S1. C, tandem-affinity purification of His6-biotinylation domain-His6-tagged KH (HBH::KH), followed by tandem mass tagging and tandem MS identification. Results of two replicate experiments comparing parasites expressing or not expressing HBH::KH are shown, and the proteins with the greatest differential in tandem-mass-tag intensities common to both replicates listed as in B. This analysis excluded ribosomal proteins and one endogenously biotinylated protein, as these are common contaminant proteins and are unlikely to be true KH interaction partners. More detailed experimental data are reported in Tables S2 and S3.