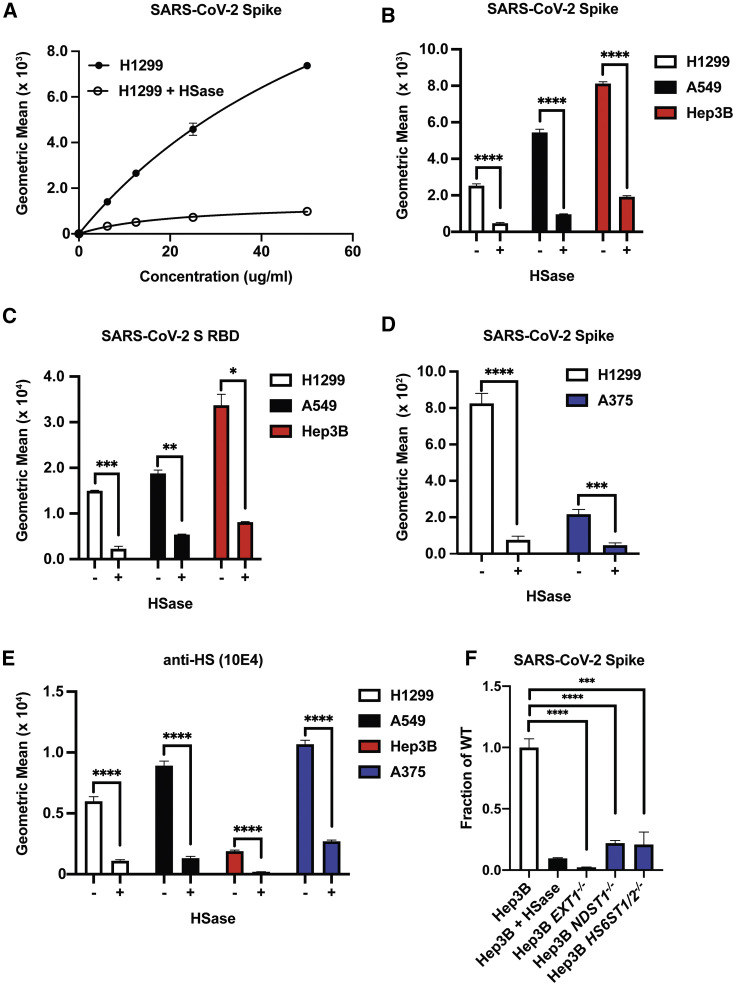
Figure 3.
SARS-CoV-2 Spike Ectodomain Binding to Cells Is Dependent on Cellular HS
(A) Titration of recombinant SARS-CoV-2 spike protein binding to human H1299 cells with and without treatment with a mix of heparin lyases I, II, and III (HSase).
(B) Recombinant SARS-CoV-2 spike protein binding (20 μg/mL) to H1299, A549, and Hep3B cells with and without HSase treatment.
(C) SARS-CoV-2 S RBD protein binding (20 μg/mL) to H1299, A549, and Hep3B cells with and without HSase treatment.
(D) SARS-CoV-2 spike protein binding (20 μg/mL) to H1299 and A375 cells with and without HSase treatment.
(E) Anti-HS (F58-10E4) staining of H1299, A549, Hep3B, and A375 cells with and without HSase treatment.
(F) Binding of recombinant SARS-CoV-2 spike protein (20 μg/mL) to Hep3B mutants altered in HS biosynthesis enzymes. Specific enzymes that were lacking in the mutants are listed along the x axis. All values were obtained by flow cytometry. Graphs shows representative experiments performed in technical triplicate. The experiments were repeated at least three times. Statistical analysis by unpaired t test (ns: p > 0.05, ∗: p ≤ 0.05, ∗∗: p ≤ 0.01, ∗∗∗: p ≤ 0.001, ∗∗∗∗: p ≤ 0.0001).
See also Figure S4.