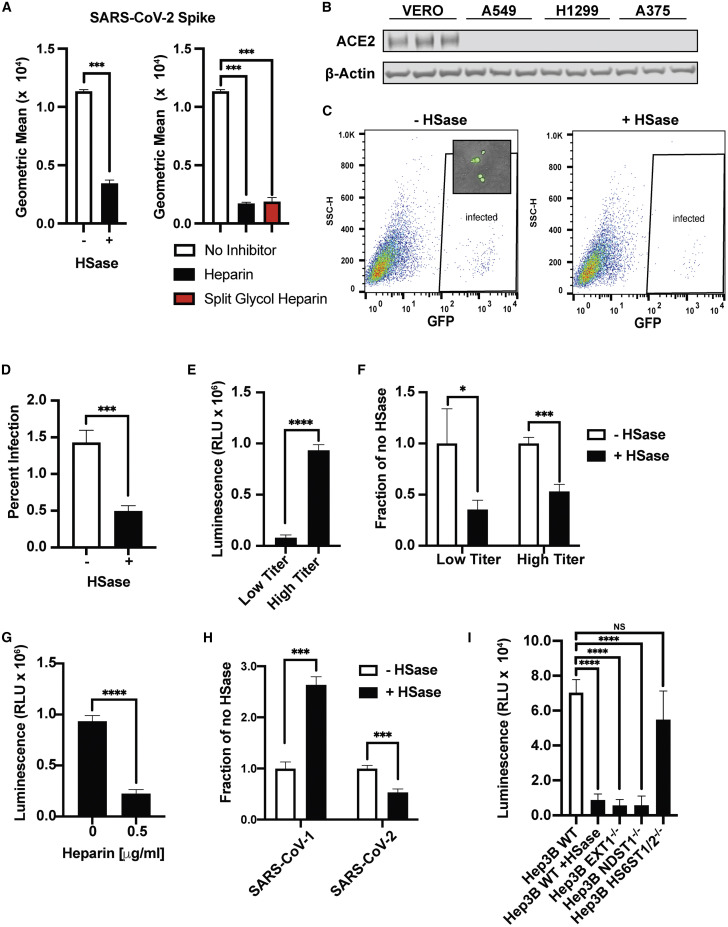
Figure 6.
SARS-CoV-2 Pseudovirus Infection Depends on Heparan Sulfate
(A) Left, SARS-CoV-2 spike protein (20 μg/mL) binding to Vero cells measured by flow cytometry with and without HSase. Right, heparin and split-glycol heparin inhibit SARS-CoV-2 spike protein (20 μg/mL) binding to Vero cells by flow cytometry. Statistical analysis by unpaired t test.
(B) Western blot analysis of ACE2 expression in Vero E6 cells compared to A549, H1299, and A375 cells. A representative blot of three extracts is shown for each strain.
(C) Infection of Vero E6 cells with SARS-CoV-2 spike protein expressing pseudotyped virus expressing GFP. Infection was done with and without HSase treatment of the cells. Insert shows GFP expression in the infected cells by imaging. Counting was performed by flow cytometry with gating for GFP-positive cells as indicated by “infected.”
(D) Quantitative analysis of GFP-positive cells.
(E) Infection of Vero E6 cells with SARS-CoV-2 S protein pseudotyped virus expressing luciferase, as measured by the addition of Bright-Glo and detection of luminescence. The figure shows infection experiments done at low and high titer.
(F) HSase treatment diminishes infection by SARS-CoV-2 S protein pseudotyped virus (luciferase) at low and high titer.
(G) Heparin (0.5 μg/mL) blocks infection with SARS-CoV-2 S protein pseudotyped virus (luciferase).
(H) Effect of HSase treatment of Vero E6 cells on the infection of both SARS-CoV-1 S and SARS-CoV-2 S protein pseudotyped virus expressing luciferase.
(I) Infection of Hep3B with and without HSase and in Hep3B cells containing mutations in EXT1, NDST1, and HS6ST1/HS6ST2. Cells were infected with SARS-CoV-2 S protein pseudotyped virus expressing luciferase. All experiments were repeated at least three times. Graphs shows representative experiments performed in technical triplicates. Statistical analysis by unpaired t test. (ns: p > 0.05, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001).
See also Figure S6.