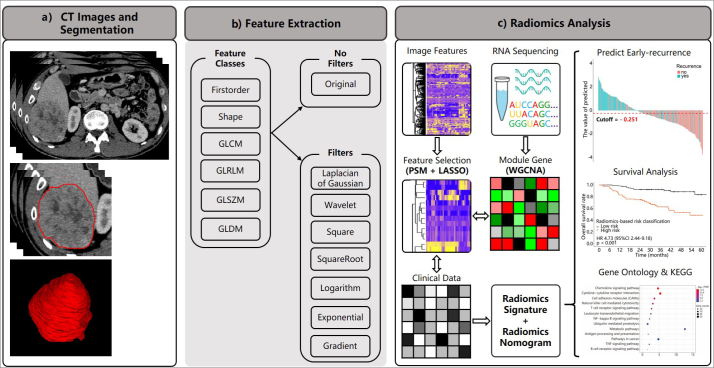
Figure 1. a–c.
The workflow of the current study. In panel (a), we obtained venous-phase contrast-enhanced CT images and segmented the volume of interest; panel (b) shows categories of extracted features; panel (c) shows the workflow outlining the process of features extraction and selection, of radiomics signatures and radiomics nomogram building, clinical utility assessment of the model (prediction of early recurrence and survival risk stratification), an exploratory analysis of small sample to construct the gene module with weighted gene co-expression network analysis (WGCNA), and the association between the predictive value of radiomics features and gene pathways using gene ontology and the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases.