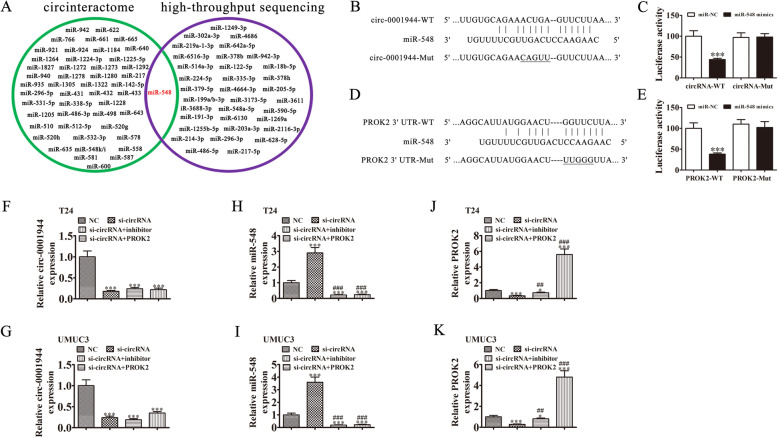
Fig. 5.
miR-548 and PROK2 are downstream targets of hsa_circ_0001944. a Venn diagram from Interactome (https://circinteractome.nia.nih.gov/), and high-throughput sequencing of miRNAs predicted to be sponged by hsa_circ_0001944. b Bioinformatics analysis prediction of binding sites of miR-548 in hsa_circ_0001944. The mutant version of hsa_circ_0001944 is presented. c Relative luciferase activity determined 48 h after transfecting HEK293T cells with miR-548 mimic/NC or hsa_circ_0001944 wild-type/Mut. Data are presented as means ± SD. ***P < 0.001. d Prediction of binding sites of miR-548 within the 3′-UTR of PROK2. The mutant version of the 3′-UTR-PROK2 is shown. e Relative luciferase activity 48 h after transfecting HEK293T cells with miR-548 mimic/NC or 3′-UTR-PROK2 wild-type/Mut. Data are presented as means ± SD. ***P < 0.001. f-k RT-qPCR detection show the expression of hsa_circ_0001944 (F and G), miR-548 (H and I), and PROK2 (J and K) in both T24 and UMUC3 cells. Data are presented as means ± SD. *P < 0.05, ***P < 0.001 vs NC. ##P < 0.01, ###P < 0.001 vs si-circRNA