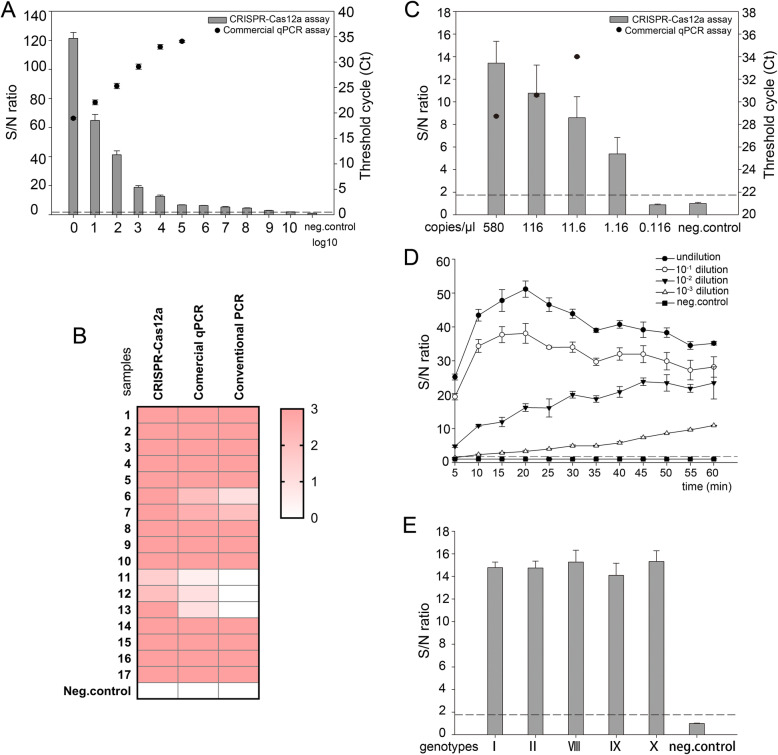
Fig. 3.
Sensitivity of CRISPR-Cas12a based assay in the detection of ASFV. a A qPCR positive blood sample (GD/GZ/0311) was subjected to serial (log10) dilution and examined by both the CRISPR-Cas12a and the commercial qPCR assay. Columns – CRISPR-Cas12a based assay; black dot – commercial qPCR assay. The dashed line represents the detection threshold for the virus. b A total of 17 qPCR positive blood were used for comparison using all three nucleic acid detection methods. Heatmap results represent the signal intensity of individual samples. c ASFV B646L Gene Plasmid Reference Material (5.8 × 103 copies/μl) was subjected to serial dilution and examined by the CRISPR-Cas12a and the commercial qPCR. Columns – CRISPR-Cas12a based assay; black dot – commercial qPCR assay. Dashed line represents the detection threshold for the virus. d The panel shows that serial dilution of the positive sample (GD/GZ/0311) remained detectable over a 1 h time course with S/N ratio (fluorescence) measurements every 5 min. e The p72 genes of various ASFV genotypes I, II, VIII, IX, and X were synthesized and used to assess whether the CRISPR-Cas12a assay has the ability to test different ASFV genotypes. The dashed line represents the detection threshold for the virus. n = 3 technical replicates, bars represent mean ± SD