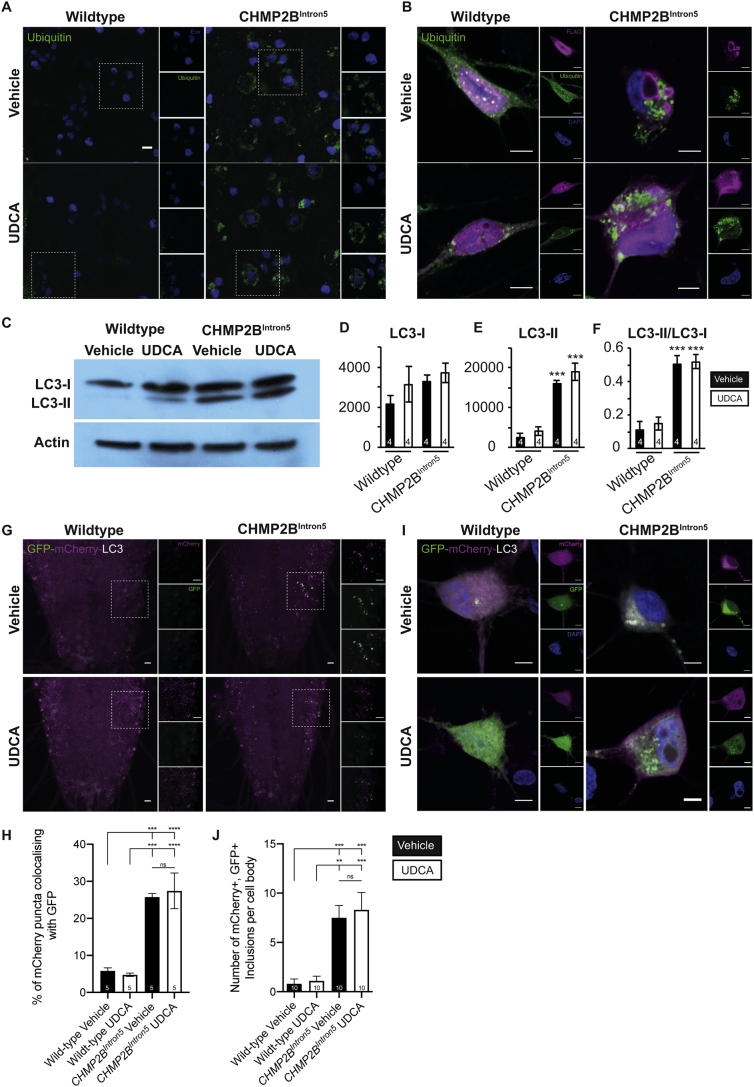
Fig. 4.
UDCA Does Not Rescue Accumulation of Ubiquitin-positive Aggregates and Autophagosomes In Drosophila and Mammalian Neurons Expressing CHMP2BIntron5.
A. Ubiquitinated proteins in the CNS of wild type and CHMP2BIntron5 expressing (nSyb-Gal4) Drosophila third instar larvae raised on vehicle (ethanol) or UDCA (600 μM) supplemented food. Drosophila motor neurons are labelled using an eve-eGFP endogenous reporter (blue). Scale bars = 5 μm.
B. Representative micrographs of mature neurons expressing FLAG-tagged CHMP2BWildtype or CHMP2BIntron5 (FLAG, magenta) ± UDCA (10 μM, 48 h) stained for ubiquitinated (green) proteins. Nuclei counterstained with DAPI (blue). Scale bar = 5 μm.
C—F. Quantification of LC3-I and LC3-II in the Drosophila larval CNS. Immunoblots (C) from 4 independent biological replicates were quantified for cytosolic LC3-I (D) and LC3-phosphatidylethanolamine conjugate (LC3-II) (E), which is recruited to autophagosomal membranes. F. LC3-I/LC3-II ratio. n = 4. All Normalised against actin loading. ANOVA with post-hoc Dunnett's comparison to wild type controls ⁎⁎⁎p < .001, ⁎⁎⁎⁎p < .0001.
G, H. Autophagic flux in the CNS of wild type and CHMP2BIntron5 expressing (nSyb-Gal4) Drosophila third instar larvae raised on vehicle (ethanol) or UDCA (600 μM) supplemented food. The dual tagged GFP-mCherry-LC3/Atg8a labels autophagosomes (GFP and mCherry), amphisomes (mCherry only) and autolysosomes (mCherry only). Scale bars = 10 μm. Quantification (H) of the percentage of LC3 aggregates positive for both mCherry and GFP in the Drosophila larval nervous system. ANOVA with post-hoc Tukey's comparison between groups ⁎⁎⁎p < .001, ⁎⁎⁎⁎p < .0001. Quantification from 5 independent animals per condition (N = 5).
I. Representative micrographs of mature neurons expressing FLAG-tagged CHMP2BWildtype or CHMP2BIntron5 (FLAG, magenta), co-transfected with a plasmid encoding GFP-mCherry-LC3. Cells were treated with vehicle (0.1% ethanol) or UDCA (10 μM, 48 h) and stained for GFP (green) and mCherry (red). Autophagosome accumulation was monitored by observing the presence of green and red puncta. Nuclei counterstained with DAPI (blue). Scale bar = 5 μm.
J. Quantification of micrographs in (I) showing the number of GFP and mCherry double positive aggregates per cell. n = 10 cells per condition. ANOVA with post-hoc Tukey's comparison between groups ⁎⁎⁎p < .001, ⁎⁎p < .01. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)