Figure 6.
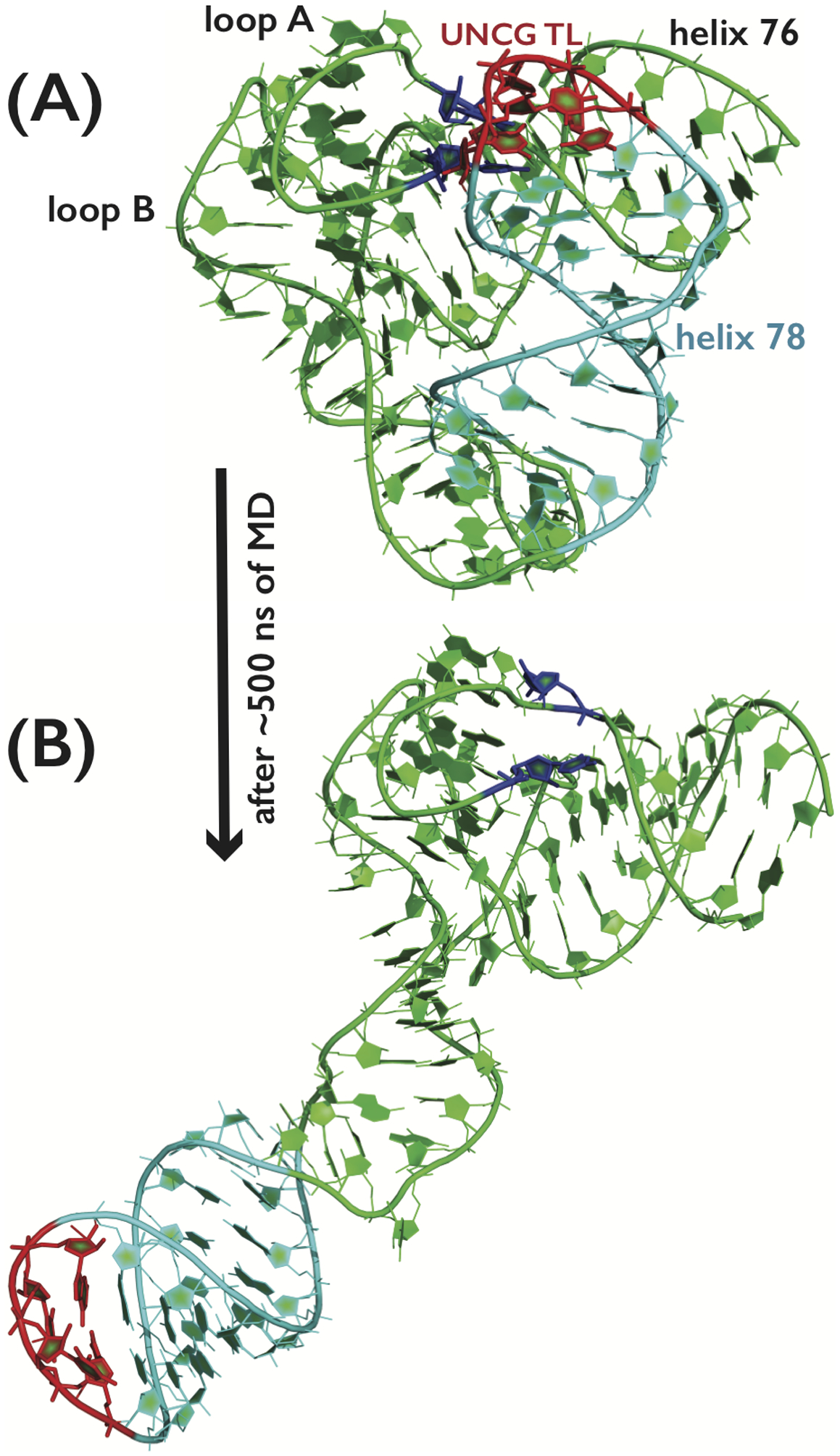
The example of poor description of the folded RNA molecule containing several noncanonical interactions during classical MD simulations with the recent DESRES60 RNA ff. (A) The starting snapshot from the MD simulation of the ribosomal L1-stalk RNA segment, where the compact fold is characterized by presence of two Kink-turns and by numerous tertiary interactions between three RNA loops, including the UNCG TL (highlighted in red; bases forming base-pair triplets with UNCG TL nucleotides are highlighted in blue). Water molecules, counter ions and hydrogen atoms are not shown for clarity. (B) Last snapshot from the simulation of the same system, where loss of several tertiary interactions, coupled with poor description of Kink-turns, was followed by large-scale degradation of the RNA fold. The degradation began immediately and fully progressed till the end of MD. Basically the same behavior was observed in a number of independent simulations (see Supporting Information for details).
