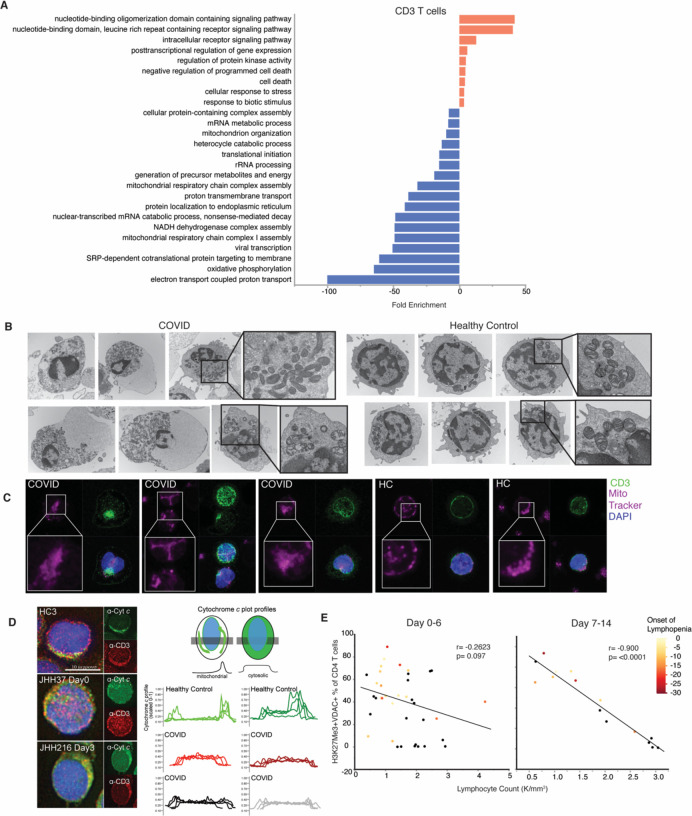
Figure 3: T cells from COVID-19 subjects demonstrate parameters of mitochondrial dysfunction and apoptosis signaling, correlating with development of lymphopenia.
A. Single-cell RNA sequencing analysis of 6 COVID-19 subjects and 3 HC were evaluated for CD3+ T cells. Genes distinguishing T cells from COVID-19 patients compared to HC were evaluated for statistical over representation using GO biological processes as gene sets and categorized into higher level annotation using ReviGO. Displayed is the enrichment score for each gene set and color corresponds to programs in upregulated genes (red) and downregulated genes (blue). B. Representative electron microscopy images of PBMCs from a COVID-A patient and a healthy control. C. Representative confocal images of PBMCs from a COVID-A patient and healthy control with mitochondria labeled using MitoTracker Deep Red (pink), CD3+ T cells labeled (green) and nuclei labeled with DAPI (blue). D. Representative fluorescence images of PBMCs from 3 COVID-A subjects and one healthy control (left) immunostained for cytochrome c (green) and CD3 (red), and nuclei labeled with DAPI (blue). Plot profiles of intracellular cytochrome c fluorescence intensity distribution (right). E. Correlation of lymphocyte count with frequency of H3K27Me3+VDAC+ out of total CD4+ T cells from day 0–6 (left) or day 7–14 (right) of study enrollment, including all symptomatic patients. Each dot represents one subject and color corresponds to days since onset of lymphopenia (black = no COVID-19 related lymphopenia developed). Correlation tested using non-parametic Spearman correlation.