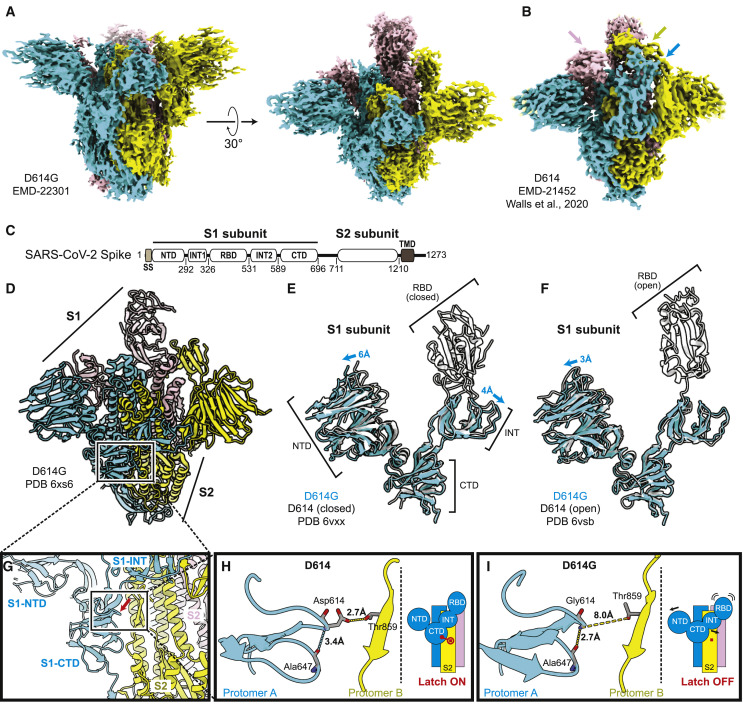
Figure 6.
Structural Determination of Spike D614G
(A) D614G envelope from the three-dimensional reconstruction. EMDB: EMD-22301.
(B) Published D614 envelope (EMD-21452). Arrows point to the density corresponding to the receptor-binding domains, which is missing in the corresponding positions in (A).
(C) Domain arrangement of the SARS-CoV-2 S protein.
(D) Atomic model for D614G without the receptor-binding domain. PDB: 6XS6.
(E and F) Comparison of the D614G S1 subunit with the closed conformation (E) and open conformation (F) of the D614 S1 subunit. Arrows indicate the relative movement of the S1 subunit of D614G.
(G) Position of amino acid 614 on the S protein.
(H and I) Substitution of Asp614 with glycine changes hydrogen bonding around residue 614. In the case of D614 (H), an inter-protomer hydrogen bond is detected. For D614G (I), the Asp614-Thr859 hydrogen bond is eliminated, and interaction with intradomain Ala647 is strengthened.
See also Figures S1 and S2.