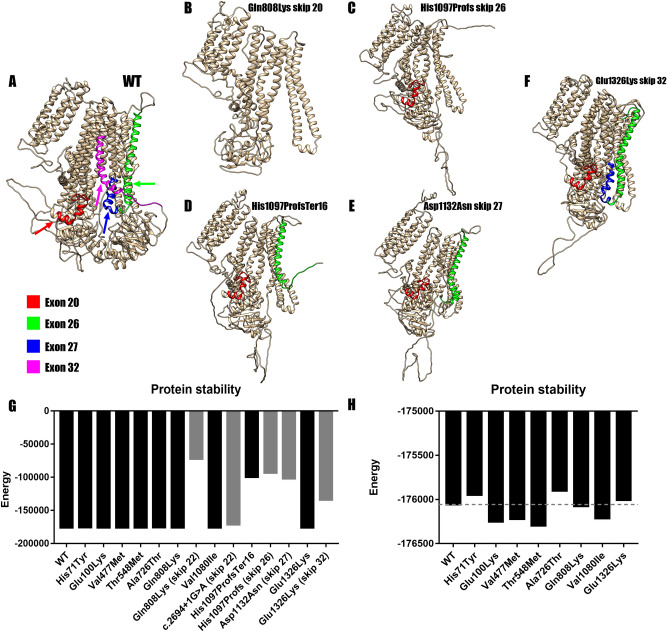
Figure 3.
Protein modeling of the most unstable variants. (A) Wild type SUR1 marked for the target exon. Red marks exon 20, green for exon 26, blue for the exon 27 and magenta for exon 32. (B) Structure for Gln808Lys in the case of an exon skipping event. The resulting protein loses 747 amino acids and so its structure is altered. (C) Structure of His1097ProfsTer16 skipping exon 26. The protein loses 527 amino acids altering its structure. (D) Structure of the resulting protein of His1097ProfsTer16. The protein loses a total of 471 amino acids and its structure is altered. (E) Structure after the mRNA confirmed skipping of exon 27 due to p.(Asp1132Asn). The skipping of exon 27 truncates the protein making it lose 463 amino acids and altering its structure. (F) Structure for Glu1326Lys after the skipping of exon 32. The protein loses 290 amino acids; the structure is altered but not as much as in earlier skips. (G) Comparison of DOPE energies between the models of the wild type (WT) SUR1 and the mutated proteins (Black for missense models and gray for exon skippings). All the amino acid substitutions showed similar stability when compared to the wild type. However, Gln808Lys (exon 22), His1097ProfsTer16, His1097ProfsTer16 (skip 26), Asp1132Asn (skip 27) and Glu1326Lys (skip 32) showed a much lower stability which would mean more unstable proteins. (H) Detailed graphic comparing all the missense variants and the WT template. None of the variants showed high DOPE energy variation when compared with the WT.