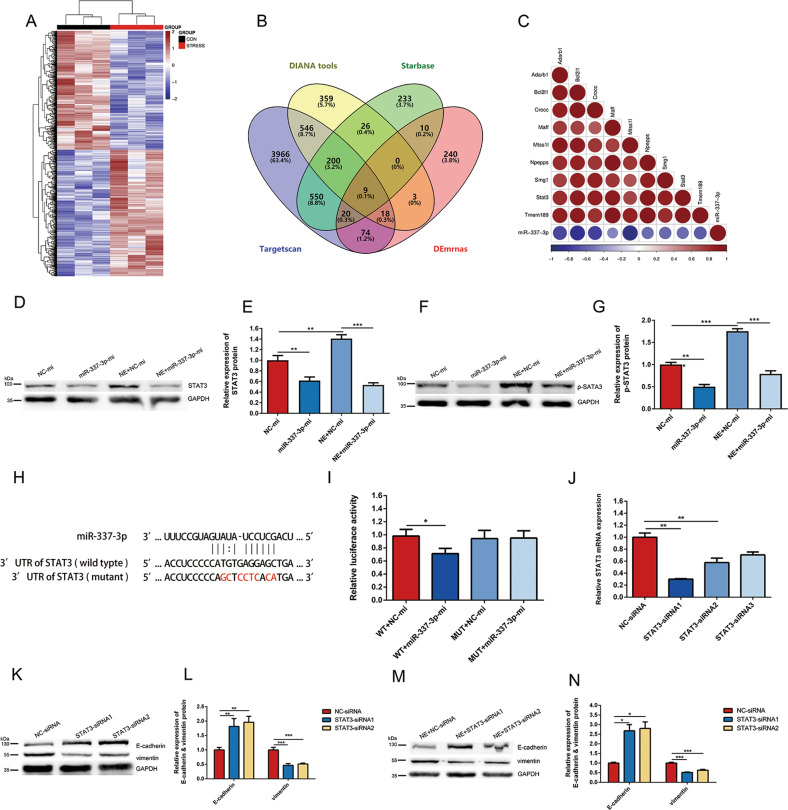
Fig. 4. miR-337-3p inhibited EMT of 4T1 cells by suppressing STAT3.
a Heat map showing significantly changed (P < 0.05, two-tailed Student’s t-test) mRNAs between control group and stress group. b A venn diagram between the differentially expressed mRNAs in a and miR-337-3p target genes from three databases (DIANA tools, Starbase, and Target scan) revealed nine potential targets. c Correlation heat map showing relationship between the 9 mRNAs in Fig. 4b and miR-337-3p. d–g miR-337-3p-mi or NC-mi were transfected into 4T1 cells cultured with or without NE. After 48 h, the expression levels of STAT3 and p-STAT3 were detected by western blot (n = 3). The intensity of the bands for STAT3 and p-STAT3 was normalized to GAPDH and shown in histogram. h The structure and sequence of the miRNA: target interactions for miR-337-3p and the 3′UTRs of STAT3, and the mutant sites in the 3′UTRs of STAT3 is highlighted in red. i Luciferase activity assays was performed to confirm the direct binding efficiency of miR-337-3p and its putative target STAT3 (n = 3). j The transfection efficiency of the knockdown of STAT3 by siRNAs in 4T1 cells was detected by RT-qPCR analysis (n = 3). Relative mRNA levels were determined by the ΔΔCt method using GAPDH for internal cross-normalization. k–n 4T1 cells were transfected with 800 ng/ml STAT3-siRNA1, STAT3-siRNA2, or NC-siRNA and the expression of E-cadherin and vimentin were detected by western blot (n = 3). The intensity of the bands for E-cadherin and vimentin was normalized to GAPDH and shown in diagram. Data are shown as the mean ± SD; *P < 0.05; **P < 0.01; ***P < 0.001.