Fig. 2. LET1 functions genetically downstream of MEKK2 and upstream of SUMM2 in cell death control.
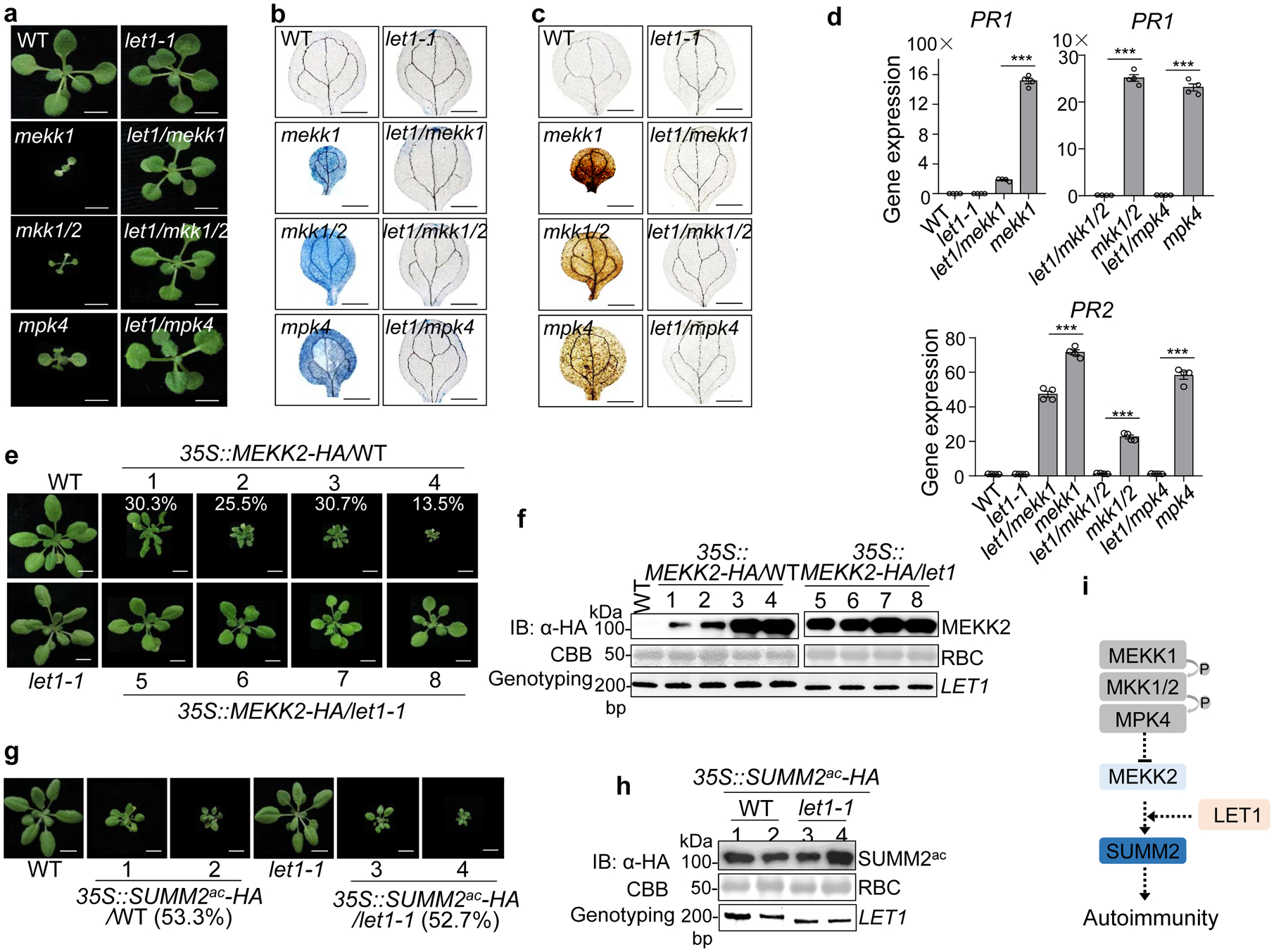
a, The let1–1 mutant suppresses growth defects of mekk1, mkk1/2, and mpk4 mutants. Three-week-old plants grown on ½ MS medium plates with indicated genotypes are shown. Scale bar, 0.5 cm.
b and c The let1–1 mutant suppresses cell death (b) and H2O2 accumulation (c) in mekk1, mkk1/2, and mpk4. Cell death and H2O2 were stained by trypan blue and DAB respectively, with cotyledons of two-week-old plants grown on ½ MS medium plates. Scale bar, 0.1 cm.
d, The let1–1 mutant suppresses the elevated expression levels of PR1 and PR2 in mekk1, mkk1/2, and mpk4. The expression of PR1 and PR2 was normalized by the expression of UBQ10. P = 3.37 × 10−8 (PR1, left, column 3 and 4), P = 3.81 × 10−8 (PR1, right, column 1 and 2), P = 8.32 × 10−8 (PR1, right, column 3 and 4), P = 4.21 × 10−5 (PR2, column 3 and 4), P = 1.21 × 10−6 (PR2, column 5 and 6), P = 2.22 × 10−7 (PR2, column 7 and 8). Data are shown as mean ± SE from four independent repeats. The asterisks indicate statistical significance by using two-sided two-tailed Student’s t-test (***, P < 0.001).
e, The let1–1 mutant suppresses growth defects triggered by overexpressing MEKK2. The 35S::MEKK2-HA transgenic plants in the WT background were grouped into four categories with representative plants showing dwarfism. The numbers are the percentages of plants displaying the indicated phenotype among a total of 251 transgenic plants analyzed for 35S::MEKK2-HA/WT (n=251). The 35S::MEKK2-HA transgenic plants in the let1–1 mutant background do not show severe growth defects and cell death (n=56). Scale bar, 1 cm.
f, MEKK2 protein expression and genotyping of LET1 in transgenic plants from e. Total proteins were subjected to immunoblotting using an α-HA antibody (top panel), and CBB staining serves as a loading control (middle panel). PCR genotyping of LET1 with primers spanning the deletion site in let1–1 is shown (bottom panel).
g, The let1–1 mutant cannot suppress the growth defects triggered by overexpressing the active SUMM2 (SUMM2ac), D478V variant. The 35S::SUMM2ac-HA transgenic plants show growth defects with a similar ratio in WT (53.3%, n=75) and let1–1 (52.7%, n=55) backgrounds. Scale bar, 1 cm.
h, SUMM2ac protein expression and genotyping of LET1 in transgenic plants from g. Assays were done similarly as in f.
i, The schematic diagram of LET1 in the SUMM2-mediated cell death pathway.
All the above experiments were repeated at least three times with similar results obtained.
