Fig. 3. MEKK2 stabilizes LET1 and SUMM2 and promotes cell death.
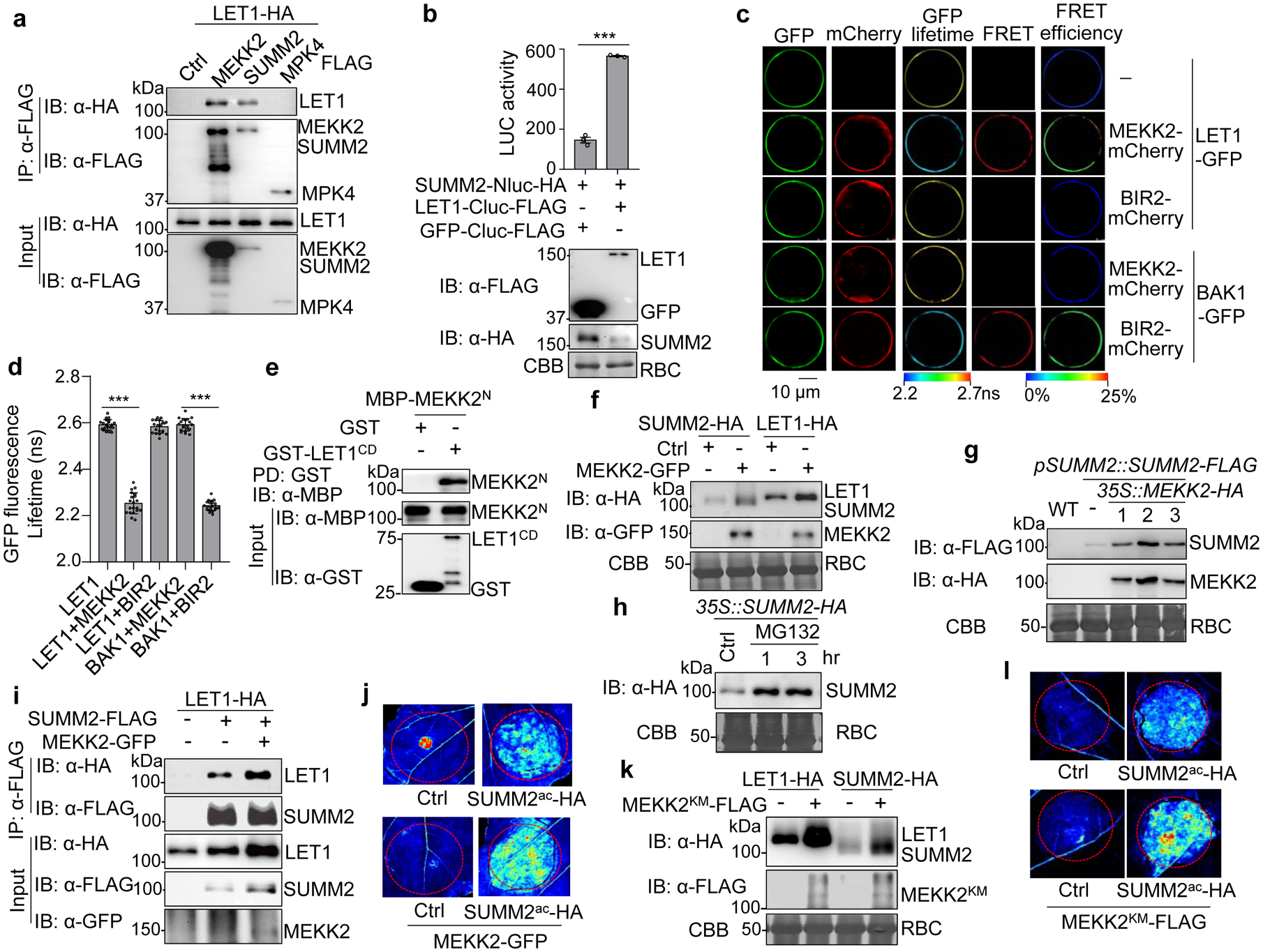
a, LET1 associates with MEKK2 and SUMM2, but not MPK4, with a Co-IP assay. LET1-HA was co-expressed with a vector control (Ctrl), MEKK2-FLAG, SUMM2-FLAG, or MPK4-FLAG, in WT Col-0 protoplasts. Total proteins were immunoprecipitated with α-FLAG affinity beads and then immunoblotted by an α-HA or α-FLAG antibody (top two panels). Proteins before immunoprecipitation are shown as input controls (bottom two panels).
b, LET1 associates with SUMM2 with a split-luciferase assay. The LET1-Cluc-FLAG or GFP-Cluc-FLAG (Ctrl) was co-expressed with SUMM2-Nluc-HA in protoplasts, and luciferase activity was detected 12 hr after transfection, P = 9.00 × 10−6. Protein expression is shown on the bottom with immunoblots. Data are shown as mean ± SE from three independent repeats. The asterisk indicates statistical significance by using two-sided two-tailed Student’s t-test (***, P<0.001).
c and d, FRET-FLIM analysis of LET1 and MEKK2 interaction in Arabidopsis protoplasts. The indicated proteins were transiently expressed in protoplasts for 16 hr, and FRET-FLIM was visualized using a confocal laser scanning microscopy (c). Localization of the LET1-GFP/BAK1-GFP and MEKK2-mCherry/BIR2-mCherry is shown with the first (Green) and second column (Red), respectively. The lifetime (τ) distribution (third column), FRET (fourth column), and apparent FRET efficiency (fifth column) are presented as pseudo-color images according to the scale. The GFP mean fluorescence lifetime (τ) values, ranging from 2.2 to 2.7 nanoseconds (ns), were statistically analyzed and data are shown as mean ± SE from twenty independent cells (d), P <1.00 × 10−15 (column 1 and 2), P <1.00 × 10−15 (column 4 and 5). The pairs of BAK1-GFP/MEKK2-mCherry and LET1-GFP/BIR2-mCherry were used as a negative control, and BAK1-GFP and BIR2-mCherry as a positive control53. Asterisks represent significant differences by using two-sided two-tailed Student’s t-test (***, P < 0.001). Scale bar, 10 μm.
e, LET1CD interacts direcstly with the N-terminal MEKK2 (MEKK2N). LET1CD consists of the juxtamembrane and kinase domains (425–834aa). MBP-MEKK2N and GST-LET1CD proteins purified from E. coli were used in an in vitro pull-down assay using glutathione agarose beads followed by immunoblotting using an α-MBP antibody (top panel). Input proteins were immunoblotted by an α-MBP or α-GST antibody before pull-down (middle and bottom panels).
f, MEKK2 enhances the LET1 and SUMM2 protein levels. LET1-HA or SUMM2-HA was co-expressed with a vector control (Ctrl) or MEKK2-GFP in N. benthamiana. Total proteins were isolated and immunoblotted by an α-HA or α-GFP antibody two days after infiltration. CBB was used for a loading control.
g, MEKK2 enhances SUMM2 protein accumulation in transgenic plants. The pMDC-35S::MEKK2-HA construct was transformed into pSUMM2::SUMM2-FLAG transgenic plants. SUMM2 and MEKK2 protein levels were examined by α-FLAG and α-HA antibodies in T1 generation of pSUMM2::SUMM2-FLAG/35S::MEKK2-HA transgenic plants (Line #1, #2, and #3). The SUMM2 protein level in the parental pSUMM2::SUMM2-FLAG line was used as a control (lane 2). The protein loading is shown by CBB staining.
h, MG132 increases SUMM2 protein level. Transgenic plants of 35S::SUMM2-HA in WT were treated with DMSO (Ctrl) or 2 μM MG132 for 1 or 3 hr. Proteins were immunoblotted using an α-HA antibody (top panel), and CBB was used for a loading control (bottom panel).
i, MEKK2 enhances the association of LET1 and SUMM2. LET1-HA was co-expressed with a vector (−), or SUMM2-FLAG with or without MEKK2-GFP in N. benthamiana and followed by a Co-IP assay.
j, MEKK2 enhances the cell death triggered by SUMM2ac. SUMM2ac-HA was co-expressed with or without MEKK2-GFP in N. benthamiana. The GFP construct was used as a Ctrl. The images were taken at two days after infiltration under the UV light.
k, The MEKK2 kinase mutant (MEKK2KM) enhances the LET1 and SUMM2 protein levels. The experiment was performed similarly as in f.
l, MEKK2KM enhances the cell death triggered by SUMM2ac. The experiment was performed similarly as in j.
All the above experiments were repeated at least three times with similar results obtained.
