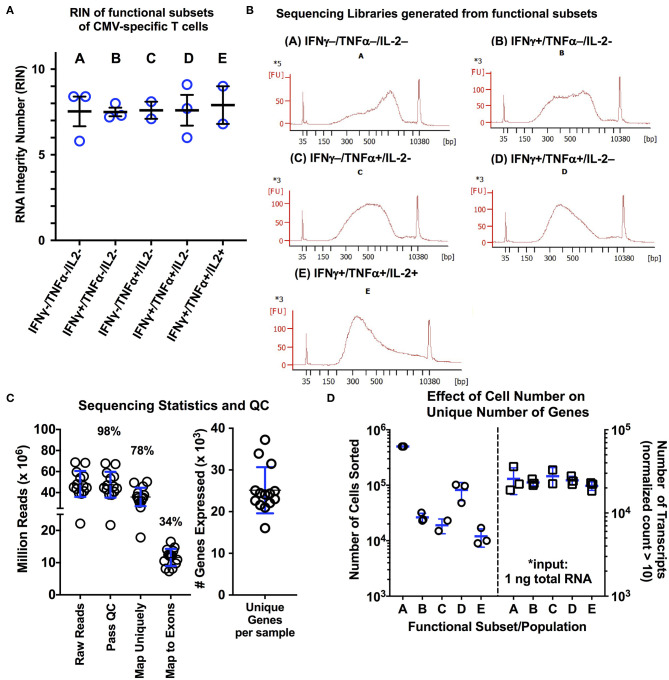
Figure 2.
(A,B) RNA quality measurements and representative cDNA libraries for functional subsets CMV-specific CD8+ T cells isolated using the modified protocol. Following activation with a CMV peptide pool, cells were processed and stained using the modified protocol, and the five predominant functional subsets of CMV-specific CD8+ T cells were sorted. Following RNA isolation using the Ambion FFPE Recoverall kit, RIN values were obtained for three different donors (n = 3 subjects; blue lines on graph represent mean ± SD). Of the 15 individual RNA samples, two of the RNA electropherograms were unable to be fitted by the RIN algorithm due to the low overall RNA input. The average RIN value of the remaining 13 libraries was 7.60 ± 1.03. (B) Representative cDNA libraries generated using the Clontech PICO input stranded total RNA kit are shown. (C) Sequencing statistics and quality control analysis results for the 15 libraries following sequencing on the Illumina HiSeq4000 platform (2 x 150 bp; 20% PhiX spike-in; 5 samples per lane). The average number of unique transcripts detected was 25.127 ± 5.53 (x103) per sample (blue lines on graph represent mean ± SD). (D) Examination of the effect of the number of sorted cells on number of unique transcripts, as a measure of RNA complexity (n = 3; blue lines on graph represent mean ± SD). A total of 1 ng of total RNA was used for library generation for each population, independent of the number of cells collected during sorting. Overall, we observed no significant effect of cell number isolated during sorting on the number of unique transcripts. Statistical testing: *p < 0.05. Populations: (A) non-functional (IFNγ-/TNFα-/IL-2-); (B) IFNγ only/monofunctional (IFNγ+/TNFα-/IL-2-); (C) TNFα monofunctional (IFNγ-/TNFα+/IL-2-); (D) bifunctional (IFNγ+/TNFα+/IL-2-); and (E) polyfunctional (IFNγ+/TNFα+/IL-2+).