Figure 4.
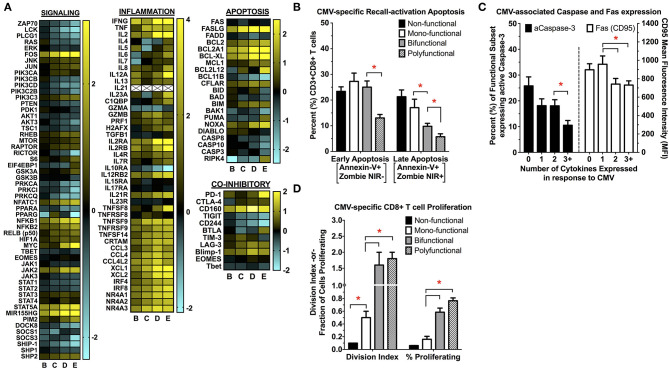
(A) Changes in cellular signaling, inflammation, apoptosis/cell survival, and co-inhibitory receptor transcript expression between the functional subsets of CMV-specific CD8+ T cells. First, genes differentially expressed between the functional subsets of CMV-specific CD8+ T cells were identified using DESeq2. This list of differentially expressed genes was then used for further in silico analysis, which identified multiple KEGG signaling pathways that were significantly upregulated in polyfunctional vs. mono- or non-functional T cells. This identified pathways involved in TCR- and cytokine-mediated signaling, viral response, inflammation, apoptosis/cell survival, and metabolism. Genes involved in these significantly-regulated pathways were then used to generate the heatmaps shown in (A) to highlight specific transcriptional patterns across the subsets (n = 5). All values are shown as log2 fold-change relative to the non-functional T cell population (pop A). The changes in inflammatory signaling seen in polyfunctional T cells is consistent with prior studies examining antigen-specific polyfunctional T cells, identified by the expression of IFNγ alone, without further functional sub-setting to specifically examining polyfunctional cells (65–68). These conserved pathways include tumor necrosis factor receptor (TNFR) signaling, effector/cytokine expression (e.g., GZMB), chemokine signaling (e.g., XCL1/2), and NR4A nuclear receptor family signaling. (B–D) Determination of CMV-specific CD8+ T cell re-activation induced apoptosis (B), expression of caspase-3 and FAS (CD95) (C), and proliferation (D) following antigen re-challenge (n = 5; error bars represent SEM). Apoptosis (early and late), caspase-3, and Fas are expressed as the percentage (%) of the parent population (functional subset), whereas proliferation is expressed as the division index (DI) and the percent (%) parent population. Statistical testing: *p < 0.05. Populations: (A) non-functional (IFNγ-/TNFα-/IL-2-); (B) IFNγ only/monofunctional (IFNγ+/TNFα-/IL-2-); (C) TNFα monofunctional (IFNγ-/TNFα+/IL-2-); (D) bifunctional (IFNγ+/TNFα+/IL-2-); and (E) polyfunctional (IFNγ+/TNFα+/IL-2+).